

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Glucocorticoid receptor | ||
| Ligand | BDBM50340664 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_739803 (CHEMBL1762863) | ||
| Ki | 2±n/a nM | ||
| Citation |  Hudson, AR; Higuchi, RI; Roach, SL; Valdez, LJ; Adams, ME; Vassar, A; Rungta, D; Syka, PM; Mais, DE; Marschke, KB; Zhi, L Nonsteroidal 2,3-dihydroquinoline glucocorticoid receptor agonists with reduced PEPCK activation. Bioorg Med Chem Lett21:1654-7 (2011) [PubMed] Article Hudson, AR; Higuchi, RI; Roach, SL; Valdez, LJ; Adams, ME; Vassar, A; Rungta, D; Syka, PM; Mais, DE; Marschke, KB; Zhi, L Nonsteroidal 2,3-dihydroquinoline glucocorticoid receptor agonists with reduced PEPCK activation. Bioorg Med Chem Lett21:1654-7 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Glucocorticoid receptor | |||
| Name: | Glucocorticoid receptor | ||
| Synonyms: | GCR_HUMAN | GR | GRL | Glucocorticoid | Glucocorticoid receptor (GRFP) | NR3C1 | Nuclear receptor subfamily 3 group C member 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 85656.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P04150 | ||
| Residue: | 777 | ||
| Sequence: |
| ||
| BDBM50340664 | |||
| n/a | |||
| Name | BDBM50340664 | ||
| Synonyms: | 6-(3-chloro-1H-indol-7-yl)-5,7-difluoro-2,2-dimethyl-2,3-dihydroquinolin-4(1H)-one O-tert-butyl oxime | CHEMBL1762203 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H24ClF2N3O | ||
| Mol. Mass. | 431.906 | ||
| SMILES | CC(C)(C)O\N=C1/CC(C)(C)Nc2cc(F)c(c(F)c12)-c1cccc2c(Cl)c[nH]c12 |(33.41,-19.38,;32.08,-20.14,;30.75,-19.36,;32.06,-18.6,;32.07,-21.68,;30.73,-22.44,;30.72,-23.97,;32.06,-24.76,;32.04,-26.3,;33.36,-25.52,;33.36,-27.06,;30.7,-27.06,;29.38,-26.28,;28.05,-27.03,;26.73,-26.26,;25.39,-27.03,;26.73,-24.73,;28.06,-23.96,;28.06,-22.42,;29.39,-24.74,;25.41,-23.95,;25.41,-22.41,;24.07,-21.63,;22.74,-22.41,;22.73,-23.95,;21.58,-24.98,;20.25,-24.21,;22.21,-26.39,;23.75,-26.23,;24.07,-24.72,)| | ||
| Structure | 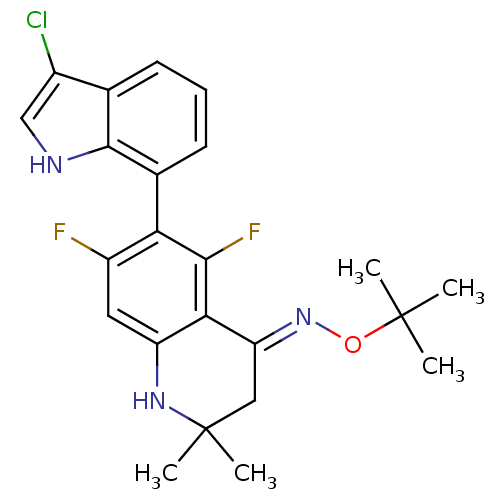
| ||