| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Uracil-DNA glycosylase |
|---|
| Ligand | BDBM50343395 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_747132 (CHEMBL1777498) |
|---|
| IC50 | 34100±n/a nM |
|---|
| Citation |  Nuth, M; Huang, L; Saw, YL; Schormann, N; Chattopadhyay, D; Ricciardi, RP Identification of inhibitors that block vaccinia virus infection by targeting the DNA synthesis processivity factor D4. J Med Chem54:3260-7 (2011) [PubMed] Article Nuth, M; Huang, L; Saw, YL; Schormann, N; Chattopadhyay, D; Ricciardi, RP Identification of inhibitors that block vaccinia virus infection by targeting the DNA synthesis processivity factor D4. J Med Chem54:3260-7 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Uracil-DNA glycosylase |
|---|
| Name: | Uracil-DNA glycosylase |
|---|
| Synonyms: | OPG116 | UDG | UNG | UNG_VACCW |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 25071.48 |
|---|
| Organism: | Vaccinia virus (strain Western Reserve) (VACV) (Vaccinia virus (strainWR)) |
|---|
| Description: | ChEMBL_747132 |
|---|
| Residue: | 218 |
|---|
| Sequence: | MNSVTVSHAPYTITYHDDWEPVMSQLVEFYNEVASWLLRDETSPIPDKFFIQLKQPLRNK
RVCVCGIDPYPKDGTGVPFESPNFTKKSIKEIASSISRLTGVIDYKGYNLNIIDGVIPWN
YYLSCKLGETKSHAIYWDKISKLLLQHITKHVSVLYCLGKTDYSNIRAKLESPVTTIVGY
HPAARDRQFEKDRSFEIINVLLELDNKAPINWAQGFIY
|
|
|
|---|
| BDBM50343395 |
|---|
| n/a |
|---|
| Name | BDBM50343395 |
|---|
| Synonyms: | 10-(cyclohexylamino)-12-[(2-hydroxyethyl)amino]-15-oxa-14-azatetracyclohexadeca-1(16),2,4,6,9,11,13-heptaen-8-one | CHEMBL1774936 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H25N3O3 |
|---|
| Mol. Mass. | 379.4522 |
|---|
| SMILES | OCCN=C1CC(NC2CCCCC2)c2c(O)c3ccccc3c3onc1c23 |w:3.2| |
|---|
| Structure | 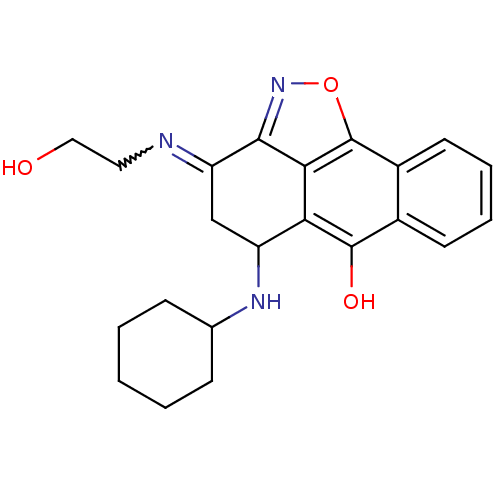
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nuth, M; Huang, L; Saw, YL; Schormann, N; Chattopadhyay, D; Ricciardi, RP Identification of inhibitors that block vaccinia virus infection by targeting the DNA synthesis processivity factor D4. J Med Chem54:3260-7 (2011) [PubMed] Article
Nuth, M; Huang, L; Saw, YL; Schormann, N; Chattopadhyay, D; Ricciardi, RP Identification of inhibitors that block vaccinia virus infection by targeting the DNA synthesis processivity factor D4. J Med Chem54:3260-7 (2011) [PubMed] Article