| Citation |  Jones, CK; Engers, DW; Thompson, AD; Field, JR; Blobaum, AL; Lindsley, SR; Zhou, Y; Gogliotti, RD; Jadhav, S; Zamorano, R; Bogenpohl, J; Smith, Y; Morrison, R; Daniels, JS; Weaver, CD; Conn, PJ; Lindsley, CW; Niswender, CM; Hopkins, CR Discovery, synthesis, and structure-activity relationship development of a series of N-4-(2,5-dioxopyrrolidin-1-yl)phenylpicolinamides (VU0400195, ML182): characterization of a novel positive allosteric modulator of the metabotropic glutamate receptor 4 (mGlu(4)) with oral efficacy in an antiparkin J Med Chem54:7639-47 (2011) [PubMed] Article Jones, CK; Engers, DW; Thompson, AD; Field, JR; Blobaum, AL; Lindsley, SR; Zhou, Y; Gogliotti, RD; Jadhav, S; Zamorano, R; Bogenpohl, J; Smith, Y; Morrison, R; Daniels, JS; Weaver, CD; Conn, PJ; Lindsley, CW; Niswender, CM; Hopkins, CR Discovery, synthesis, and structure-activity relationship development of a series of N-4-(2,5-dioxopyrrolidin-1-yl)phenylpicolinamides (VU0400195, ML182): characterization of a novel positive allosteric modulator of the metabotropic glutamate receptor 4 (mGlu(4)) with oral efficacy in an antiparkin J Med Chem54:7639-47 (2011) [PubMed] Article |
|---|
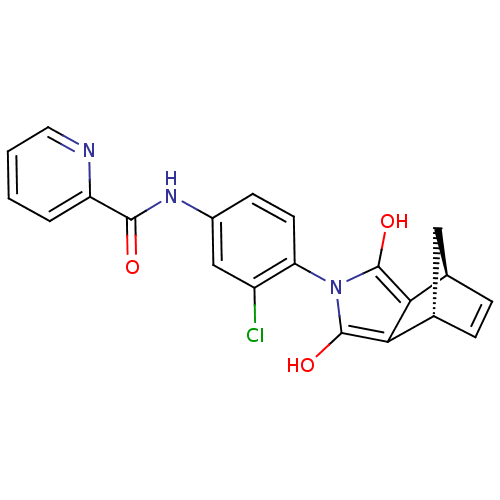
| SMILES | Oc1c2[C@H]3C[C@H](C=C3)c2c(O)n1-c1ccc(NC(=O)c2ccccn2)cc1Cl |r,wU:3.3,5.4,c:6,(-.76,-35.66,;.38,-36.71,;1.89,-36.4,;2.67,-35.09,;2.65,-36.63,;4.17,-37.76,;4.95,-36.44,;4.2,-35.1,;2.64,-37.74,;1.61,-38.88,;1.92,-40.39,;.21,-38.24,;-1.11,-39.04,;-2.45,-38.29,;-3.77,-39.09,;-3.75,-40.63,;-5.07,-41.42,;-6.42,-40.67,;-6.44,-39.13,;-7.74,-41.46,;-7.71,-43,;-9.03,-43.8,;-10.38,-43.05,;-10.4,-41.51,;-9.08,-40.72,;-2.4,-41.38,;-1.08,-40.59,;.27,-41.33,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jones, CK; Engers, DW; Thompson, AD; Field, JR; Blobaum, AL; Lindsley, SR; Zhou, Y; Gogliotti, RD; Jadhav, S; Zamorano, R; Bogenpohl, J; Smith, Y; Morrison, R; Daniels, JS; Weaver, CD; Conn, PJ; Lindsley, CW; Niswender, CM; Hopkins, CR Discovery, synthesis, and structure-activity relationship development of a series of N-4-(2,5-dioxopyrrolidin-1-yl)phenylpicolinamides (VU0400195, ML182): characterization of a novel positive allosteric modulator of the metabotropic glutamate receptor 4 (mGlu(4)) with oral efficacy in an antiparkin J Med Chem54:7639-47 (2011) [PubMed] Article
Jones, CK; Engers, DW; Thompson, AD; Field, JR; Blobaum, AL; Lindsley, SR; Zhou, Y; Gogliotti, RD; Jadhav, S; Zamorano, R; Bogenpohl, J; Smith, Y; Morrison, R; Daniels, JS; Weaver, CD; Conn, PJ; Lindsley, CW; Niswender, CM; Hopkins, CR Discovery, synthesis, and structure-activity relationship development of a series of N-4-(2,5-dioxopyrrolidin-1-yl)phenylpicolinamides (VU0400195, ML182): characterization of a novel positive allosteric modulator of the metabotropic glutamate receptor 4 (mGlu(4)) with oral efficacy in an antiparkin J Med Chem54:7639-47 (2011) [PubMed] Article