

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Heat shock protein HSP 90-beta | ||
| Ligand | BDBM50361700 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_798935 (CHEMBL1943442) | ||
| IC50 | 8.8±n/a nM | ||
| Citation |  Baruchello, R; Simoni, D; Grisolia, G; Barbato, G; Marchetti, P; Rondanin, R; Mangiola, S; Giannini, G; Brunetti, T; Alloatti, D; Gallo, G; Ciacci, A; Vesci, L; Castorina, M; Milazzo, FM; Cervoni, ML; Guglielmi, MB; Barbarino, M; Foderà, R; Pisano, C; Cabri, W Novel 3,4-isoxazolediamides as potent inhibitors of chaperone heat shock protein 90. J Med Chem54:8592-604 (2011) [PubMed] Article Baruchello, R; Simoni, D; Grisolia, G; Barbato, G; Marchetti, P; Rondanin, R; Mangiola, S; Giannini, G; Brunetti, T; Alloatti, D; Gallo, G; Ciacci, A; Vesci, L; Castorina, M; Milazzo, FM; Cervoni, ML; Guglielmi, MB; Barbarino, M; Foderà, R; Pisano, C; Cabri, W Novel 3,4-isoxazolediamides as potent inhibitors of chaperone heat shock protein 90. J Med Chem54:8592-604 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Heat shock protein HSP 90-beta | |||
| Name: | Heat shock protein HSP 90-beta | ||
| Synonyms: | HS90B_HUMAN | HSP 84 | HSP 90 | HSP90AB1 | HSP90B | HSPC2 | HSPCB | Heat Shock Protein 90 (Hsp90) | Heat shock protein HSP 90 (Hsp90) | ||
| Type: | Molecular Chaperone | ||
| Mol. Mass.: | 83229.45 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08238 | ||
| Residue: | 724 | ||
| Sequence: |
| ||
| BDBM50361700 | |||
| n/a | |||
| Name | BDBM50361700 | ||
| Synonyms: | CHEMBL1941049 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H38N4O6 | ||
| Mol. Mass. | 514.6138 | ||
| SMILES | CCNC(=O)c1noc(c1NC(=O)[C@H]1CC[C@H](CN2CCOCC2)CC1)-c1cc(C(C)C)c(O)cc1O |r,wU:13.13,wD:16.17,(25.27,-47.09,;23.73,-47.08,;22.96,-45.74,;21.42,-45.74,;20.76,-44.35,;20.55,-47.01,;21.07,-48.46,;19.85,-49.4,;18.57,-48.52,;19.01,-47.05,;18.07,-45.83,;16.53,-45.8,;15.74,-47.11,;15.79,-44.45,;14.26,-44.42,;13.52,-43.07,;14.32,-41.75,;13.59,-40.4,;12.05,-40.36,;11.31,-39.01,;9.78,-38.97,;8.97,-40.28,;9.7,-41.64,;11.25,-41.68,;15.86,-41.79,;16.6,-43.14,;17.24,-49.3,;15.91,-48.53,;14.58,-49.3,;13.25,-48.53,;13.25,-46.99,;11.91,-49.3,;14.58,-50.85,;13.24,-51.62,;15.91,-51.62,;17.25,-50.85,;18.59,-51.61,)| | ||
| Structure | 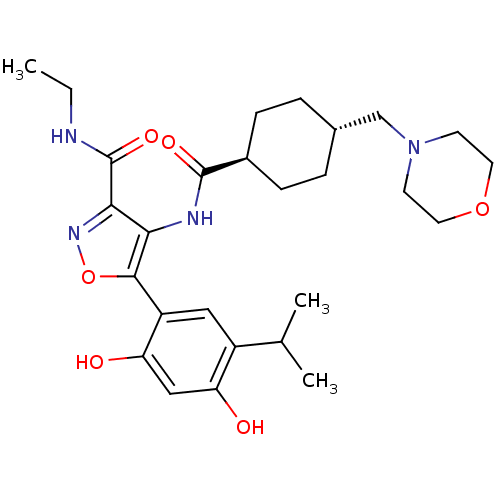
| ||