

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M2 | ||
| Ligand | BDBM50072228 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_139656 | ||
| EC50 | 62±n/a nM | ||
| Citation |  Sauerberg, P; Jeppesen, L; Olesen, PH; Sheardown, MJ; Fink-Jensen, A; Rasmussen, T; Rimvall, K; Shannon, HE; Bymaster, FP; DeLapp, NW; Calligaro, DO; Ward, JS; Whitesitt, CA; Thomsen, C Identification of side chains on 1,2,5-thiadiazole-azacycles optimal for muscarinic m1 receptor activation. Bioorg Med Chem Lett8:2897-902 (1999) [PubMed] Sauerberg, P; Jeppesen, L; Olesen, PH; Sheardown, MJ; Fink-Jensen, A; Rasmussen, T; Rimvall, K; Shannon, HE; Bymaster, FP; DeLapp, NW; Calligaro, DO; Ward, JS; Whitesitt, CA; Thomsen, C Identification of side chains on 1,2,5-thiadiazole-azacycles optimal for muscarinic m1 receptor activation. Bioorg Med Chem Lett8:2897-902 (1999) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M2 | |||
| Name: | Muscarinic acetylcholine receptor M2 | ||
| Synonyms: | ACM2_HUMAN | CHRM2 | Cholinergic, muscarinic M2 | Muscarinic acetylcholine receptor M2 and M4 | Muscarinic acetylcholine receptor M2 and M5 | RecName: Full=Muscarinic acetylcholine receptor M2 | ||
| Type: | GPCR | ||
| Mol. Mass.: | 51730.61 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08172 | ||
| Residue: | 466 | ||
| Sequence: |
| ||
| BDBM50072228 | |||
| n/a | |||
| Name | BDBM50072228 | ||
| Synonyms: | 3-[4-(3-Phenyl-prop-2-ynyloxy)-[1,2,5]thiadiazol-3-yl]-1-aza-bicyclo[2.2.2]octane | CHEMBL99521 | NNC 11-1314 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H19N3OS | ||
| Mol. Mass. | 325.428 | ||
| SMILES | C(Oc1nsnc1C1CN2CCC1CC2)C#Cc1ccccc1 |(14.79,-5.45,;13.26,-5.32,;12.37,-6.6,;12.89,-8.07,;11.66,-9,;10.38,-8.12,;10.82,-6.64,;9.88,-5.4,;8.44,-5.96,;7.22,-5.01,;8.49,-3.99,;8.96,-4.86,;10.1,-3.86,;8.91,-2.91,;7.44,-3.72,;15.47,-6.86,;16.24,-8.18,;16.99,-9.52,;18.54,-9.52,;19.29,-10.85,;18.51,-12.2,;16.98,-12.2,;16.22,-10.85,)| | ||
| Structure | 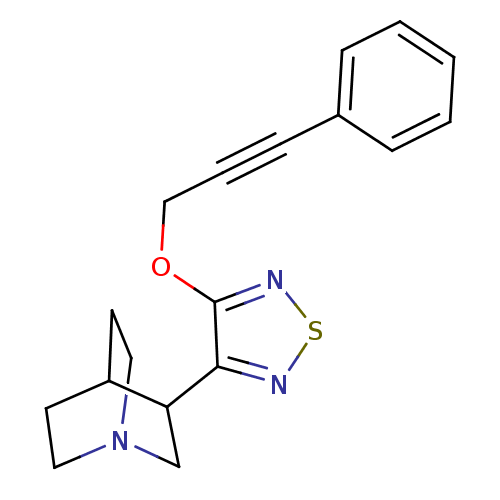
| ||