

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cholecystokinin receptor type A | ||
| Ligand | BDBM50061999 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_50191 (CHEMBL663484) | ||
| IC50 | 2239±n/a nM | ||
| Citation |  Trivedi, BK; Padia, JK; Holmes, A; Rose, S; Wright, DS; Hinton, JP; Pritchard, MC; Eden, JM; Kneen, C; Webdale, L; Suman-Chauhan, N; Boden, P; Singh, L; Field, MJ; Hill, D Second generation"peptoid" CCK-B receptor antagonists: identification and development of N-(adamantyloxycarbonyl)-alpha-methyl-(R)-tryptophan derivative (CI-1015) with an improved pharmacokinetic profile. J Med Chem41:38-45 (1998) [PubMed] Article Trivedi, BK; Padia, JK; Holmes, A; Rose, S; Wright, DS; Hinton, JP; Pritchard, MC; Eden, JM; Kneen, C; Webdale, L; Suman-Chauhan, N; Boden, P; Singh, L; Field, MJ; Hill, D Second generation"peptoid" CCK-B receptor antagonists: identification and development of N-(adamantyloxycarbonyl)-alpha-methyl-(R)-tryptophan derivative (CI-1015) with an improved pharmacokinetic profile. J Med Chem41:38-45 (1998) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cholecystokinin receptor type A | |||
| Name: | Cholecystokinin receptor type A | ||
| Synonyms: | CCKAR_RAT | Cckar | Cholecystokinin peripheral | Cholecystokinin receptor | Cholecystokinin receptor type A | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 49676.37 | ||
| Organism: | RAT | ||
| Description: | Cholecystokinin central 0 RAT::P30551 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50061999 | |||
| n/a | |||
| Name | BDBM50061999 | ||
| Synonyms: | CHEMBL138952 | [(R)-1-(1-Benzyl-2-phenyl-ethylcarbamoyl)-2-(1H-indol-3-yl)-1-methyl-ethyl]-carbamic acid adamantan-2-yl ester | ||
| Type | Small organic molecule | ||
| Emp. Form. | C38H43N3O3 | ||
| Mol. Mass. | 589.7663 | ||
| SMILES | C[C@](Cc1c[nH]c2ccccc12)(NC(=O)OC1C2CC3CC(C2)CC1C3)C(=O)NC(Cc1ccccc1)Cc1ccccc1 |wU:1.13,wD:1.0,TLB:25:24:22:19.18.20,THB:25:19:16.24.23:22,20:19:16:21.23.22,20:21:16:19.18.25,15:16:22:19.18.20,(-.04,-9,;-.02,-10.36,;.05,-11.9,;1.43,-12.61,;.17,-13.52,;.65,-14.99,;2.2,-14.98,;3.21,-16.13,;4.72,-15.81,;5.2,-14.34,;4.17,-13.21,;2.67,-13.52,;-1.42,-9.85,;-2.7,-10.7,;-2.68,-12.16,;-4.08,-10.01,;-5.37,-10.87,;-5.38,-12.4,;-6.78,-12.75,;-8.1,-12.26,;-9.3,-13.53,;-7.8,-13.11,;-6.4,-13.68,;-7.81,-11.52,;-6.77,-10.29,;-8.13,-10.77,;1.25,-9.68,;1.24,-8.3,;2.63,-10.38,;3.91,-9.52,;5.29,-10.21,;6.79,-9.8,;7.16,-8.33,;8.63,-7.91,;9.73,-9,;9.35,-10.48,;7.86,-10.89,;3.82,-7.98,;5.1,-7.13,;6.48,-7.81,;7.77,-6.97,;7.67,-5.43,;6.27,-4.74,;5,-5.59,)| | ||
| Structure | 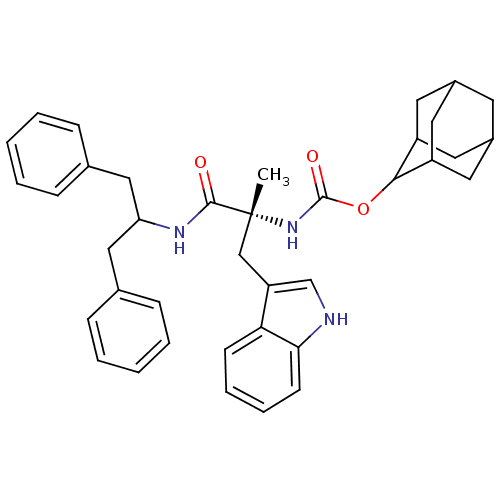
| ||