

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Reverse transcriptase/RNaseH | ||
| Ligand | BDBM50036599 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_195058 (CHEMBL797513) | ||
| IC50 | 13600±n/a nM | ||
| Citation |  Smith, RH; Jorgensen, WL; Tirado-Rives, J; Lamb, ML; Janssen, PA; Michejda, CJ; Kroeger Smith, MB Prediction of binding affinities for TIBO inhibitors of HIV-1 reverse transcriptase using Monte Carlo simulations in a linear response method. J Med Chem41:5272-86 (1999) [PubMed] Article Smith, RH; Jorgensen, WL; Tirado-Rives, J; Lamb, ML; Janssen, PA; Michejda, CJ; Kroeger Smith, MB Prediction of binding affinities for TIBO inhibitors of HIV-1 reverse transcriptase using Monte Carlo simulations in a linear response method. J Med Chem41:5272-86 (1999) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Reverse transcriptase/RNaseH | |||
| Name: | Reverse transcriptase/RNaseH | ||
| Synonyms: | HIV-1 Reverse Transcriptase RNase H | Human immunodeficiency virus type 1 reverse transcriptase | Reverse transcriptase/RNaseH | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 65229.15 | ||
| Organism: | Human immunodeficiency virus 1 | ||
| Description: | ChEMBL_1473730 | ||
| Residue: | 566 | ||
| Sequence: |
| ||
| BDBM50036599 | |||
| n/a | |||
| Name | BDBM50036599 | ||
| Synonyms: | (S)-5,8-Dimethyl-7-(3-methyl-but-2-enyl)-6,7,8,9-tetrahydro-2H-2,7,9a-triaza-benzo[cd]azulene-1-thione | 5,8-Dimethyl-7-(3-methyl-but-2-enyl)-6,7,8,9-tetrahydro-2H-2,7,9a-triaza-benzo[cd]azulene-1-thione | CHEMBL59279 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H23N3S | ||
| Mol. Mass. | 301.45 | ||
| SMILES | C[C@H]1Cn2c3c(CN1CC=C(C)C)c(C)ccc3[nH]c2=S |wD:1.0,(11.98,-15.32,;11.98,-13.78,;13.36,-13.12,;13.69,-11.61,;12.7,-10.4,;11.18,-10.41,;10.22,-11.63,;10.58,-13.12,;9.38,-14.11,;7.93,-13.56,;6.74,-14.53,;5.29,-13.98,;6.98,-16.05,;10.4,-9.08,;8.86,-9.1,;11.16,-7.75,;12.7,-7.75,;13.48,-9.08,;14.97,-9.41,;15.14,-10.93,;16.47,-11.69,)| | ||
| Structure | 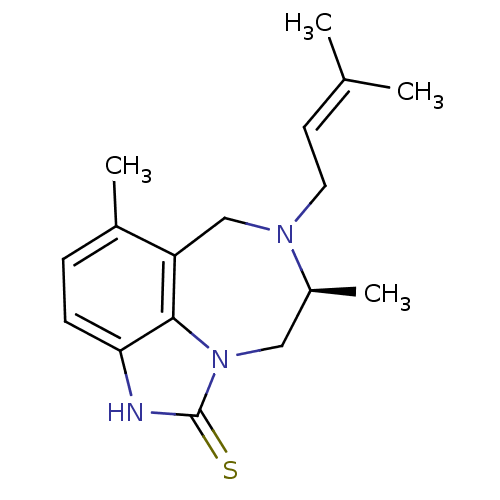
| ||