

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Polyunsaturated fatty acid lipoxygenase ALOX15 | ||
| Ligand | BDBM50221403 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_449594 (CHEMBL898689) | ||
| IC50 | >50000±n/a nM | ||
| Citation |  Deschamps, JD; Gautschi, JT; Whitman, S; Johnson, TA; Gassner, NC; Crews, P; Holman, TR Discovery of platelet-type 12-human lipoxygenase selective inhibitors by high-throughput screening of structurally diverse libraries. Bioorg Med Chem15:6900-8 (2007) [PubMed] Article Deschamps, JD; Gautschi, JT; Whitman, S; Johnson, TA; Gassner, NC; Crews, P; Holman, TR Discovery of platelet-type 12-human lipoxygenase selective inhibitors by high-throughput screening of structurally diverse libraries. Bioorg Med Chem15:6900-8 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Polyunsaturated fatty acid lipoxygenase ALOX15 | |||
| Name: | Polyunsaturated fatty acid lipoxygenase ALOX15 | ||
| Synonyms: | 15-Lipoxygenase-1 (15-LOX-1) | ALOX15 | Arachidonate 15-lipoxygenase | Arachidonate 15-lipoxygenase-1 | LOG15 | LOX15_HUMAN | Reticulocyte 15-lipoxygenase-1 | Sphingosine 1-phosphate receptor 1 (S1P1) | ||
| Type: | Protein | ||
| Mol. Mass.: | 74804.05 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 662 | ||
| Sequence: |
| ||
| BDBM50221403 | |||
| n/a | |||
| Name | BDBM50221403 | ||
| Synonyms: | CHEMBL397803 | mischellamine B | ||
| Type | Small organic molecule | ||
| Emp. Form. | C46H48N2O8 | ||
| Mol. Mass. | 756.8819 | ||
| SMILES | COc1cc(C)cc2c(cc(c(O)c12)-c1ccc2c(c(C)cc(OC)c2c1O)-c1c(O)cc(O)c2[C@@H](C)N[C@H](C)Cc12)-c1c(O)cc(O)c2[C@@H](C)N[C@H](C)Cc12 |wU:38.42,52.58,wD:49.55,35.39,(-8.29,-38.12,;-6.96,-37.34,;-6.96,-35.81,;-8.29,-35.04,;-8.29,-33.49,;-9.62,-32.73,;-6.96,-32.73,;-5.63,-33.48,;-4.3,-32.71,;-2.97,-33.47,;-2.96,-35.02,;-4.3,-35.8,;-4.29,-37.34,;-5.63,-35.03,;-1.63,-35.78,;-1.62,-37.32,;-.29,-38.08,;1.03,-37.31,;2.38,-38.07,;3.71,-37.29,;5.05,-38.06,;3.7,-35.74,;2.36,-34.99,;2.34,-33.45,;3.67,-32.66,;1.03,-35.76,;-.31,-35,;-.32,-33.47,;2.38,-39.6,;1.05,-40.38,;-.28,-39.61,;1.06,-41.92,;2.39,-42.68,;2.41,-44.22,;3.73,-41.89,;5.04,-42.65,;5.05,-44.19,;6.36,-41.88,;6.36,-40.35,;7.45,-39.24,;5.03,-39.59,;3.72,-40.36,;-4.31,-31.19,;-2.98,-30.4,;-1.64,-31.17,;-2.99,-28.87,;-4.32,-28.11,;-4.33,-26.57,;-5.65,-28.88,;-7,-28.13,;-7,-26.59,;-8.33,-28.9,;-8.31,-30.45,;-9.64,-31.22,;-6.97,-31.22,;-5.65,-30.42,)| | ||
| Structure | 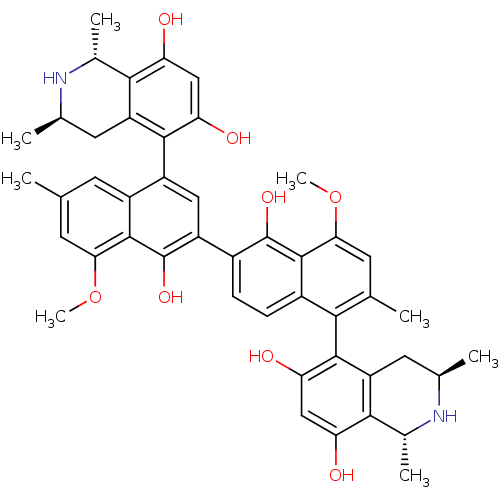
| ||