| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor 3 |
|---|
| Ligand | BDBM50373889 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_468627 (CHEMBL934444) |
|---|
| Ki | 1070±n/a nM |
|---|
| Citation |  Beich-Frandsen, M; Pickering, DS; Mirza, O; Johansen, TN; Greenwood, J; Vestergaard, B; Schousboe, A; Gajhede, M; Liljefors, T; Kastrup, JS Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency. J Med Chem51:1459-63 (2008) [PubMed] Article Beich-Frandsen, M; Pickering, DS; Mirza, O; Johansen, TN; Greenwood, J; Vestergaard, B; Schousboe, A; Gajhede, M; Liljefors, T; Kastrup, JS Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency. J Med Chem51:1459-63 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor 3 |
|---|
| Name: | Glutamate receptor 3 |
|---|
| Synonyms: | AMPA-selective glutamate receptor 3 | GLUR3 | GLURC | GRIA3 | GRIA3_HUMAN | GluR-3 | GluR-C | GluR-K3 | Glutamate receptor 3 | Glutamate receptor ionotropic AMPA | Glutamate receptor ionotropic, AMPA 3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 101172.14 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_468627 |
|---|
| Residue: | 894 |
|---|
| Sequence: | MARQKKMGQSVLRAVFFLVLGLLGHSHGGFPNTISIGGLFMRNTVQEHSAFRFAVQLYNT
NQNTTEKPFHLNYHVDHLDSSNSFSVTNAFCSQFSRGVYAIFGFYDQMSMNTLTSFCGAL
HTSFVTPSFPTDADVQFVIQMRPALKGAILSLLGHYKWEKFVYLYDTERGFSILQAIMEA
AVQNNWQVTARSVGNIKDVQEFRRIIEEMDRRQEKRYLIDCEVERINTILEQVVILGKHS
RGYHYMLANLGFTDILLERVMHGGANITGFQIVNNENPMVQQFIQRWVRLDEREFPEAKN
APLKYTSALTHDAILVIAEAFRYLRRQRVDVSRRGSAGDCLANPAVPWSQGIDIERALKM
VQVQGMTGNIQFDTYGRRTNYTIDVYEMKVSGSRKAGYWNEYERFVPFSDQQISNDSASS
ENRTIVVTTILESPYVMYKKNHEQLEGNERYEGYCVDLAYEIAKHVRIKYKLSIVGDGKY
GARDPETKIWNGMVGELVYGRADIAVAPLTITLVREEVIDFSKPFMSLGISIMIKKPQKS
KPGVFSFLDPLAYEIWMCIVFAYIGVSVVLFLVSRFSPYEWHLEDNNEEPRDPQSPPDPP
NEFGIFNSLWFSLGAFMQQGCDISPRSLSGRIVGGVWWFFTLIIISSYTANLAAFLTVER
MVSPIESAEDLAKQTEIAYGTLDSGSTKEFFRRSKIAVYEKMWSYMKSAEPSVFTKTTAD
GVARVRKSKGKFAFLLESTMNEYIEQRKPCDTMKVGGNLDSKGYGVATPKGSALRNAVNL
AVLKLNEQGLLDKLKNKWWYDKGECGSGGGDSKDKTSALSLSNVAGVFYILVGGLGLAMM
VALIEFCYKSRAESKRMKLTKNTQNFKPAPATNTQNYATYREGYNVYGTESVKI
|
|
|
|---|
| BDBM50373889 |
|---|
| n/a |
|---|
| Name | BDBM50373889 |
|---|
| Synonyms: | CHEMBL402444 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C5H7N3O3S |
|---|
| Mol. Mass. | 189.192 |
|---|
| SMILES | N[C@H](Cc1ns[nH]c1=O)C(O)=O |r| |
|---|
| Structure | 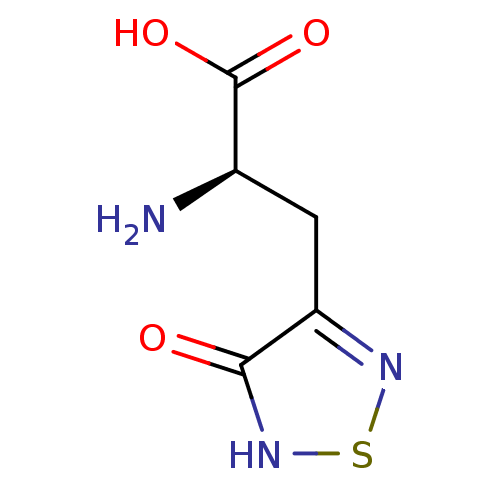
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Beich-Frandsen, M; Pickering, DS; Mirza, O; Johansen, TN; Greenwood, J; Vestergaard, B; Schousboe, A; Gajhede, M; Liljefors, T; Kastrup, JS Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency. J Med Chem51:1459-63 (2008) [PubMed] Article
Beich-Frandsen, M; Pickering, DS; Mirza, O; Johansen, TN; Greenwood, J; Vestergaard, B; Schousboe, A; Gajhede, M; Liljefors, T; Kastrup, JS Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency. J Med Chem51:1459-63 (2008) [PubMed] Article