| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase Yes |
|---|
| Ligand | BDBM50221547 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_473236 (CHEMBL932927) |
|---|
| IC50 | 4.8±n/a nM |
|---|
| Citation |  Palanki, MS; Akiyama, H; Campochiaro, P; Cao, J; Chow, CP; Dellamary, L; Doukas, J; Fine, R; Gritzen, C; Hood, JD; Hu, S; Kachi, S; Kang, X; Klebansky, B; Kousba, A; Lohse, D; Mak, CC; Martin, M; McPherson, A; Pathak, VP; Renick, J; Soll, R; Umeda, N; Yee, S; Yokoi, K; Zeng, B; Zhu, H; Noronha, G Development of prodrug 4-chloro-3-(5-methyl-3-{[4-(2-pyrrolidin-1-ylethoxy)phenyl]amino}-1,2,4-benzotriazin-7-yl)phenyl benzoate (TG100801): a topically administered therapeutic candidate in clinical trials for the treatment of age-related macular degeneration. J Med Chem51:1546-59 (2008) [PubMed] Article Palanki, MS; Akiyama, H; Campochiaro, P; Cao, J; Chow, CP; Dellamary, L; Doukas, J; Fine, R; Gritzen, C; Hood, JD; Hu, S; Kachi, S; Kang, X; Klebansky, B; Kousba, A; Lohse, D; Mak, CC; Martin, M; McPherson, A; Pathak, VP; Renick, J; Soll, R; Umeda, N; Yee, S; Yokoi, K; Zeng, B; Zhu, H; Noronha, G Development of prodrug 4-chloro-3-(5-methyl-3-{[4-(2-pyrrolidin-1-ylethoxy)phenyl]amino}-1,2,4-benzotriazin-7-yl)phenyl benzoate (TG100801): a topically administered therapeutic candidate in clinical trials for the treatment of age-related macular degeneration. J Med Chem51:1546-59 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase Yes |
|---|
| Name: | Tyrosine-protein kinase Yes |
|---|
| Synonyms: | Proto-oncogene c-Yes | VHL/Tyrosine-protein kinase Yes | YES | YES1 | YES_HUMAN | p61-yes |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 60801.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07947 |
|---|
| Residue: | 543 |
|---|
| Sequence: | MGCIKSKENKSPAIKYRPENTPEPVSTSVSHYGAEPTTVSPCPSSSAKGTAVNFSSLSMT
PFGGSSGVTPFGGASSSFSVVPSSYPAGLTGGVTIFVALYDYEARTTEDLSFKKGERFQI
INNTEGDWWEARSIATGKNGYIPSNYVAPADSIQAEEWYFGKMGRKDAERLLLNPGNQRG
IFLVRESETTKGAYSLSIRDWDEIRGDNVKHYKIRKLDNGGYYITTRAQFDTLQKLVKHY
TEHADGLCHKLTTVCPTVKPQTQGLAKDAWEIPRESLRLEVKLGQGCFGEVWMGTWNGTT
KVAIKTLKPGTMMPEAFLQEAQIMKKLRHDKLVPLYAVVSEEPIYIVTEFMSKGSLLDFL
KEGDGKYLKLPQLVDMAAQIADGMAYIERMNYIHRDLRAANILVGENLVCKIADFGLARL
IEDNEYTARQGAKFPIKWTAPEAALYGRFTIKSDVWSFGILQTELVTKGRVPYPGMVNRE
VLEQVERGYRMPCPQGCPESLHELMNLCWKKDPDERPTFEYIQSFLEDYFTATEPQYQPG
ENL
|
|
|
|---|
| BDBM50221547 |
|---|
| n/a |
|---|
| Name | BDBM50221547 |
|---|
| Synonyms: | 4-chloro-3-(5-methyl-3-(4-(3-(pyrrolidin-1-yl)propyl)phenylamino)benzo[e][1,2,4]triazin-7-yl)phenol | 7-(2-chloro-5-hydroxyphenyl)-5-methyl-N-[4-(3-pyrrolidin-1-ylpropyl)]phenyl]-1,2,4-benzotriazin-3-amine | CHEMBL250213 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H28ClN5O |
|---|
| Mol. Mass. | 473.997 |
|---|
| SMILES | Cc1cc(cc2nnc(Nc3ccc(CCCN4CCCC4)cc3)nc12)-c1cc(O)ccc1Cl |
|---|
| Structure | 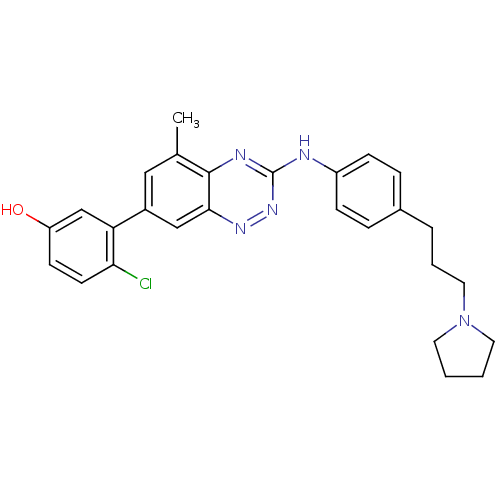
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Palanki, MS; Akiyama, H; Campochiaro, P; Cao, J; Chow, CP; Dellamary, L; Doukas, J; Fine, R; Gritzen, C; Hood, JD; Hu, S; Kachi, S; Kang, X; Klebansky, B; Kousba, A; Lohse, D; Mak, CC; Martin, M; McPherson, A; Pathak, VP; Renick, J; Soll, R; Umeda, N; Yee, S; Yokoi, K; Zeng, B; Zhu, H; Noronha, G Development of prodrug 4-chloro-3-(5-methyl-3-{[4-(2-pyrrolidin-1-ylethoxy)phenyl]amino}-1,2,4-benzotriazin-7-yl)phenyl benzoate (TG100801): a topically administered therapeutic candidate in clinical trials for the treatment of age-related macular degeneration. J Med Chem51:1546-59 (2008) [PubMed] Article
Palanki, MS; Akiyama, H; Campochiaro, P; Cao, J; Chow, CP; Dellamary, L; Doukas, J; Fine, R; Gritzen, C; Hood, JD; Hu, S; Kachi, S; Kang, X; Klebansky, B; Kousba, A; Lohse, D; Mak, CC; Martin, M; McPherson, A; Pathak, VP; Renick, J; Soll, R; Umeda, N; Yee, S; Yokoi, K; Zeng, B; Zhu, H; Noronha, G Development of prodrug 4-chloro-3-(5-methyl-3-{[4-(2-pyrrolidin-1-ylethoxy)phenyl]amino}-1,2,4-benzotriazin-7-yl)phenyl benzoate (TG100801): a topically administered therapeutic candidate in clinical trials for the treatment of age-related macular degeneration. J Med Chem51:1546-59 (2008) [PubMed] Article