| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
|---|
| Ligand | BDBM50228363 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_479068 (CHEMBL933819) |
|---|
| EC50 | >300000±n/a nM |
|---|
| Citation |  Milne, JC; Lambert, PD; Schenk, S; Carney, DP; Smith, JJ; Gagne, DJ; Jin, L; Boss, O; Perni, RB; Vu, CB; Bemis, JE; Xie, R; Disch, JS; Ng, PY; Nunes, JJ; Lynch, AV; Yang, H; Galonek, H; Israelian, K; Choy, W; Iffland, A; Lavu, S; Medvedik, O; Sinclair, DA; Olefsky, JM; Jirousek, MR; Elliott, PJ; Westphal, CH Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature450:712-716 (2007) [PubMed] Article Milne, JC; Lambert, PD; Schenk, S; Carney, DP; Smith, JJ; Gagne, DJ; Jin, L; Boss, O; Perni, RB; Vu, CB; Bemis, JE; Xie, R; Disch, JS; Ng, PY; Nunes, JJ; Lynch, AV; Yang, H; Galonek, H; Israelian, K; Choy, W; Iffland, A; Lavu, S; Medvedik, O; Sinclair, DA; Olefsky, JM; Jirousek, MR; Elliott, PJ; Westphal, CH Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature450:712-716 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
|---|
| Synonyms: | NAD-dependent deacetylase sirtuin 3 | NAD-dependent protein deacetylase sirtuin-3 (SIRT3) | SIR2L3 | SIR3_HUMAN | SIRT3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43582.69 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NTG7 |
|---|
| Residue: | 399 |
|---|
| Sequence: | MAFWGWRAAAALRLWGRVVERVEAGGGVGPFQACGCRLVLGGRDDVSAGLRGSHGARGEP
LDPARPLQRPPRPEVPRAFRRQPRAAAPSFFFSSIKGGRRSISFSVGASSVVGSGGSSDK
GKLSLQDVAELIRARACQRVVVMVGAGISTPSGIPDFRSPGSGLYSNLQQYDLPYPEAIF
ELPFFFHNPKPFFTLAKELYPGNYKPNVTHYFLRLLHDKGLLLRLYTQNIDGLERVSGIP
ASKLVEAHGTFASATCTVCQRPFPGEDIRADVMADRVPRCPVCTGVVKPDIVFFGEPLPQ
RFLLHVVDFPMADLLLILGTSLEVEPFASLTEAVRSSVPRLLINRDLVGPLAWHPRSRDV
AQLGDVVHGVESLVELLGWTEEMRDLVQRETGKLDGPDK
|
|
|
|---|
| BDBM50228363 |
|---|
| n/a |
|---|
| Name | BDBM50228363 |
|---|
| Synonyms: | CHEMBL257991 | N-(2-(3-(piperazin-1-ylmethyl)imidazo[2,1-b]thiazol-6-yl)phenyl)quinoxaline-2-carboxamide | SRT-1720 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H23N7OS |
|---|
| Mol. Mass. | 469.561 |
|---|
| SMILES | O=C(Nc1ccccc1-c1cn2c(CN3CCNCC3)csc2n1)c1cnc2ccccc2n1 |
|---|
| Structure | 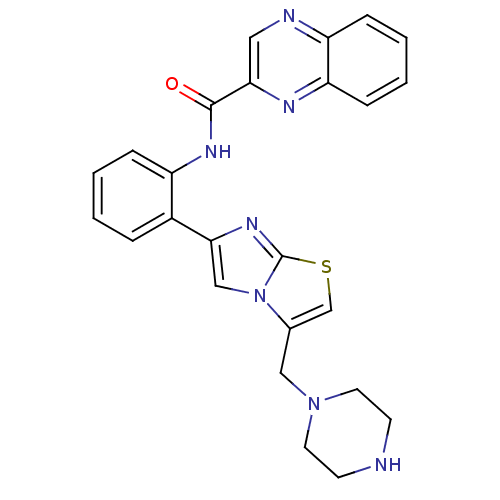
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Milne, JC; Lambert, PD; Schenk, S; Carney, DP; Smith, JJ; Gagne, DJ; Jin, L; Boss, O; Perni, RB; Vu, CB; Bemis, JE; Xie, R; Disch, JS; Ng, PY; Nunes, JJ; Lynch, AV; Yang, H; Galonek, H; Israelian, K; Choy, W; Iffland, A; Lavu, S; Medvedik, O; Sinclair, DA; Olefsky, JM; Jirousek, MR; Elliott, PJ; Westphal, CH Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature450:712-716 (2007) [PubMed] Article
Milne, JC; Lambert, PD; Schenk, S; Carney, DP; Smith, JJ; Gagne, DJ; Jin, L; Boss, O; Perni, RB; Vu, CB; Bemis, JE; Xie, R; Disch, JS; Ng, PY; Nunes, JJ; Lynch, AV; Yang, H; Galonek, H; Israelian, K; Choy, W; Iffland, A; Lavu, S; Medvedik, O; Sinclair, DA; Olefsky, JM; Jirousek, MR; Elliott, PJ; Westphal, CH Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature450:712-716 (2007) [PubMed] Article