

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 14 | ||
| Ligand | BDBM50251924 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_488009 (CHEMBL1013888) | ||
| IC50 | 15±n/a nM | ||
| Citation |  Laufer, SA; Hauser, DR; Domeyer, DM; Kinkel, K; Liedtke, AJ Design, synthesis, and biological evaluation of novel Tri- and tetrasubstituted imidazoles as highly potent and specific ATP-mimetic inhibitors of p38 MAP kinase: focus on optimized interactions with the enzyme's surface-exposed front region. J Med Chem51:4122-49 (2008) [PubMed] Article Laufer, SA; Hauser, DR; Domeyer, DM; Kinkel, K; Liedtke, AJ Design, synthesis, and biological evaluation of novel Tri- and tetrasubstituted imidazoles as highly potent and specific ATP-mimetic inhibitors of p38 MAP kinase: focus on optimized interactions with the enzyme's surface-exposed front region. J Med Chem51:4122-49 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 14 | |||
| Name: | Mitogen-activated protein kinase 14 | ||
| Synonyms: | CSAID-binding protein | CSBP | CSBP1 | CSBP2 | CSPB1 | Cytokine suppressive anti-inflammatory drug-binding protein | MAP kinase 14 | MAP kinase MXI2 | MAP kinase p38 alpha | MAPK 14 | MAPK14 | MAX-interacting protein 2 | MK14_HUMAN | MXI2 | Mitogen-activated protein kinase p38 alpha | SAPK2A | Stress-activated protein kinase 2a | p38 MAP kinase alpha/beta | ||
| Type: | Serine/threonine-protein kinase | ||
| Mol. Mass.: | 41286.76 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q16539 | ||
| Residue: | 360 | ||
| Sequence: |
| ||
| BDBM50251924 | |||
| n/a | |||
| Name | BDBM50251924 | ||
| Synonyms: | CHEMBL481751 | trans-{4-[4-(4-Fluoro-phenyl)-2-methylsulfanyl-1H-imidazol-5-yl]-pyridin-2-yl}-(4-methylcyclohexyl)-amine | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H25FN4S | ||
| Mol. Mass. | 396.524 | ||
| SMILES | CSc1nc(c([nH]1)-c1ccc(F)cc1)-c1ccnc(N[C@H]2CC[C@H](C)CC2)c1 |r,wU:20.21,wD:23.25,(2.16,-47.39,;1.39,-46.06,;-.15,-46.05,;-1.06,-47.29,;-2.52,-46.82,;-2.51,-45.27,;-1.05,-44.8,;-3.84,-44.5,;-3.83,-42.96,;-5.16,-42.18,;-6.5,-42.94,;-7.83,-42.16,;-6.5,-44.49,;-5.17,-45.26,;-3.85,-47.58,;-5.18,-46.81,;-6.52,-47.58,;-6.52,-49.12,;-5.17,-49.89,;-5.17,-51.43,;-3.83,-52.19,;-2.49,-51.42,;-1.16,-52.19,;-1.16,-53.73,;.18,-54.5,;-2.48,-54.5,;-3.81,-53.73,;-3.85,-49.11,)| | ||
| Structure | 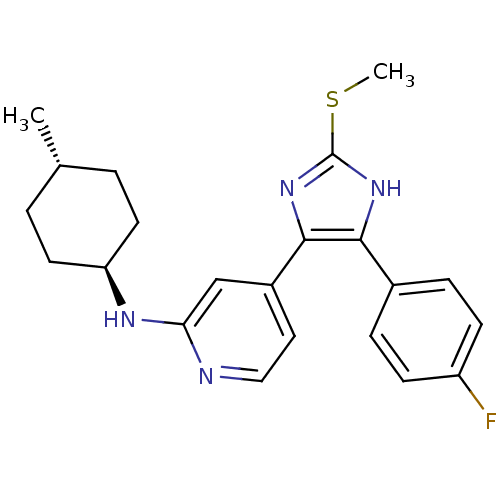
| ||