| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neuronal acetylcholine receptor subunit alpha-7 |
|---|
| Ligand | BDBM50093253 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_539612 (CHEMBL1025617) |
|---|
| Kd | 220±n/a nM |
|---|
| Citation |  Huang, X; Zheng, F; Stokes, C; Papke, RL; Zhan, CG Modeling binding modes of alpha7 nicotinic acetylcholine receptor with ligands: the roles of Gln117 and other residues of the receptor in agonist binding. J Med Chem51:6293-302 (2008) [PubMed] Article Huang, X; Zheng, F; Stokes, C; Papke, RL; Zhan, CG Modeling binding modes of alpha7 nicotinic acetylcholine receptor with ligands: the roles of Gln117 and other residues of the receptor in agonist binding. J Med Chem51:6293-302 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Neuronal acetylcholine receptor subunit alpha-7 |
|---|
| Name: | Neuronal acetylcholine receptor subunit alpha-7 |
|---|
| Synonyms: | ACHA7_HUMAN | CHRNA7 | Cholinergic, Nicotinic Alpha7 | NACHRA7 | Neuronal acetylcholine receptor protein alpha-7 subunit | Neuronal acetylcholine receptor subunit alpha-7 (nAChR-alpha 7) | Nicotinic acetylcholine receptor alpha-7 (alpha7 nAChR) |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 56448.33 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | CHRNA7 (NACHRA7) |
|---|
| Residue: | 502 |
|---|
| Sequence: | MRCSPGGVWLALAASLLHVSLQGEFQRKLYKELVKNYNPLERPVANDSQPLTVYFSLSLL
QIMDVDEKNQVLTTNIWLQMSWTDHYLQWNVSEYPGVKTVRFPDGQIWKPDILLYNSADE
RFDATFHTNVLVNSSGHCQYLPPGIFKSSCYIDVRWFPFDVQHCKLKFGSWSYGGWSLDL
QMQEADISGYIPNGEWDLVGIPGKRSERFYECCKEPYPDVTFTVTMRRRTLYYGLNLLIP
CVLISALALLVFLLPADSGEKISLGITVLLSLTVFMLLVAEIMPATSDSVPLIAQYFAST
MIIVGLSVVVTVIVLQYHHHDPDGGKMPKWTRVILLNWCAWFLRMKRPGEDKVRPACQHK
QRRCSLASVEMSAVAPPPASNGNLLYIGFRGLDGVHCVPTPDSGVVCGRMACSPTHDEHL
LHGGQPPEGDPDLAKILEEVRYIANRFRCQDESEAVCSEWKFAACVVDRLCLMAFSVFTI
ICTIGILMSAPNFVEAVSKDFA
|
|
|
|---|
| BDBM50093253 |
|---|
| n/a |
|---|
| Name | BDBM50093253 |
|---|
| Synonyms: | (+)-3'-Methylspiro[1-azabicyclo[2.2.2]octane-3,5'-oxazolidin-2'-one] | (-)-3'-Methylspiro[1-azabicyclo[2.2.2]octane-3,5'-oxazolidin-2'-one] | 3'-Methylspiro[1-azabicyclo[2.2.2]octane-3,5'-oxazolidin-2'-one] | CHEMBL129932 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H16N2O2 |
|---|
| Mol. Mass. | 196.2462 |
|---|
| SMILES | CN1CC2(CN3CCC2CC3)OC1=O |TLB:2:3:9.10:7.6,THB:11:3:9.10:7.6,(8.37,-6.07,;7.28,-4.97,;5.74,-4.97,;5.25,-3.5,;4.14,-4.08,;3.57,-2.86,;1.97,-3.38,;2.56,-2.34,;4.26,-1.93,;4.62,-.97,;3.57,-1.69,;6.51,-2.58,;7.76,-3.5,;9.23,-3.02,)| |
|---|
| Structure | 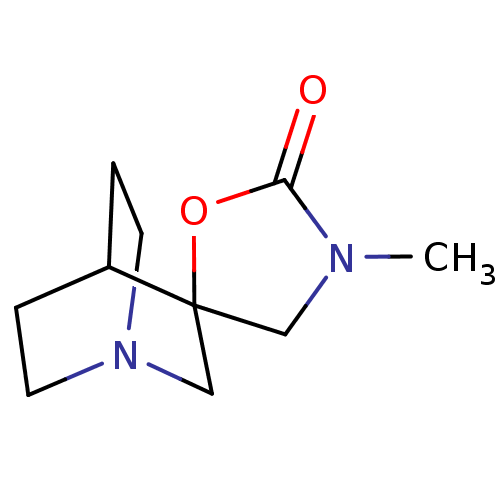
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Huang, X; Zheng, F; Stokes, C; Papke, RL; Zhan, CG Modeling binding modes of alpha7 nicotinic acetylcholine receptor with ligands: the roles of Gln117 and other residues of the receptor in agonist binding. J Med Chem51:6293-302 (2008) [PubMed] Article
Huang, X; Zheng, F; Stokes, C; Papke, RL; Zhan, CG Modeling binding modes of alpha7 nicotinic acetylcholine receptor with ligands: the roles of Gln117 and other residues of the receptor in agonist binding. J Med Chem51:6293-302 (2008) [PubMed] Article