| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dipeptidyl peptidase 2 |
|---|
| Ligand | BDBM50200723 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_657416 (CHEMBL1246392) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Tsai, TY; Yeh, TK; Chen, X; Hsu, T; Jao, YC; Huang, CH; Song, JS; Huang, YC; Chien, CH; Chiu, JH; Yen, SC; Tang, HK; Chao, YS; Jiaang, WT Substituted 4-carboxymethylpyroglutamic acid diamides as potent and selective inhibitors of fibroblast activation protein. J Med Chem53:6572-83 (2010) [PubMed] Article Tsai, TY; Yeh, TK; Chen, X; Hsu, T; Jao, YC; Huang, CH; Song, JS; Huang, YC; Chien, CH; Chiu, JH; Yen, SC; Tang, HK; Chao, YS; Jiaang, WT Substituted 4-carboxymethylpyroglutamic acid diamides as potent and selective inhibitors of fibroblast activation protein. J Med Chem53:6572-83 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dipeptidyl peptidase 2 |
|---|
| Name: | Dipeptidyl peptidase 2 |
|---|
| Synonyms: | DAP II | DPP2 | DPP2_HUMAN | DPP7 | Dipeptidyl aminopeptidase II | Dipeptidyl peptidase 2 (DPP II) | Dipeptidyl peptidase 2 (DPP2) | Dipeptidyl peptidase II (DDP-II) | Dipeptidyl peptidase II (DPP II) | Dipeptidyl peptidase II (DPP2) | Dipeptidyl peptidase II and dipeptidyl peptidase IV (DPP2 and DPP4) | QPP | carboxytripeptidase | dipeptidyl arylamidase II | dipeptidyl(amino)peptidase II | dipeptidylarylamidase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 54339.29 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UHL4 |
|---|
| Residue: | 492 |
|---|
| Sequence: | MGSAPWAPVLLLALGLRGLQAGARRAPDPGFQERFFQQRLDHFNFERFGNKTFPQRFLVS
DRFWVRGEGPIFFYTGNEGDVWAFANNSAFVAELAAERGALLVFAEHRYYGKSLPFGAQS
TQRGHTELLTVEQALADFAELLRALRRDLGAQDAPAIAFGGSYGGMLSAYLRMKYPHLVA
GALAASAPVLAVAGLGDSNQFFRDVTADFEGQSPKCTQGVREAFRQIKDLFLQGAYDTVR
WEFGTCQPLSDEKDLTQLFMFARNAFTVLAMMDYPYPTDFLGPLPANPVKVGCDRLLSEA
QRITGLRALAGLVYNASGSEHCYDIYRLYHSCADPTGCGTGPDARAWDYQACTEINLTFA
SNNVTDMFPDLPFTDELRQRYCLDTWGVWPRPDWLLTSFWGGDLRAASNIIFSNGNLDPW
AGGGIRRNLSASVIAVTIQGGAHHLDLRASHPEDPASVVEARKLEATIIGEWVKAARREQ
QPALRGGPRLSL
|
|
|
|---|
| BDBM50200723 |
|---|
| n/a |
|---|
| Name | BDBM50200723 |
|---|
| Synonyms: | (R)-1-(2-(1-oxoisoindolin-2-yl)acetyl)pyrrolidin-2-ylboronic acid | CHEMBL387050 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H17BN2O4 |
|---|
| Mol. Mass. | 288.107 |
|---|
| SMILES | OB(O)[C@@H]1CCCN1C(=O)CN1Cc2ccccc2C1=O |
|---|
| Structure | 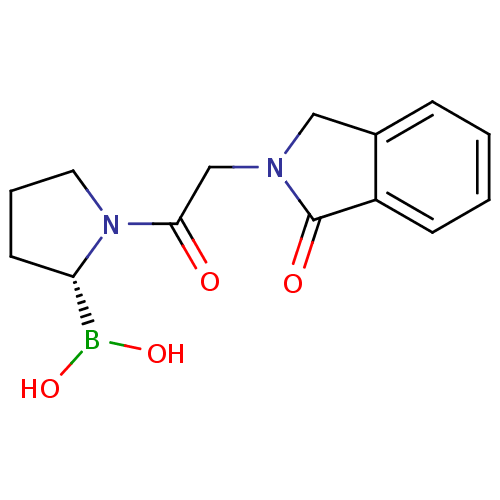
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Tsai, TY; Yeh, TK; Chen, X; Hsu, T; Jao, YC; Huang, CH; Song, JS; Huang, YC; Chien, CH; Chiu, JH; Yen, SC; Tang, HK; Chao, YS; Jiaang, WT Substituted 4-carboxymethylpyroglutamic acid diamides as potent and selective inhibitors of fibroblast activation protein. J Med Chem53:6572-83 (2010) [PubMed] Article
Tsai, TY; Yeh, TK; Chen, X; Hsu, T; Jao, YC; Huang, CH; Song, JS; Huang, YC; Chien, CH; Chiu, JH; Yen, SC; Tang, HK; Chao, YS; Jiaang, WT Substituted 4-carboxymethylpyroglutamic acid diamides as potent and selective inhibitors of fibroblast activation protein. J Med Chem53:6572-83 (2010) [PubMed] Article