| Citation |  Lainé, DI; Yan, H; Xie, H; Davis, RS; Dufour, J; Widdowson, KL; Palovich, MR; Wan, Z; Foley, JJ; Schmidt, DB; Hunsberger, GE; Burman, M; Bacon, AM; Webb, EF; Luttmann, MA; Salmon, M; Sarau, HM; Umbrecht, ST; Landis, PS; Peck, BJ; Busch-Petersen, J Design, synthesis and structure-activity relationship of N-substituted tropane muscarinic acetylcholine receptor antagonists. Bioorg Med Chem Lett22:3366-9 (2012) [PubMed] Article Lainé, DI; Yan, H; Xie, H; Davis, RS; Dufour, J; Widdowson, KL; Palovich, MR; Wan, Z; Foley, JJ; Schmidt, DB; Hunsberger, GE; Burman, M; Bacon, AM; Webb, EF; Luttmann, MA; Salmon, M; Sarau, HM; Umbrecht, ST; Landis, PS; Peck, BJ; Busch-Petersen, J Design, synthesis and structure-activity relationship of N-substituted tropane muscarinic acetylcholine receptor antagonists. Bioorg Med Chem Lett22:3366-9 (2012) [PubMed] Article |
|---|
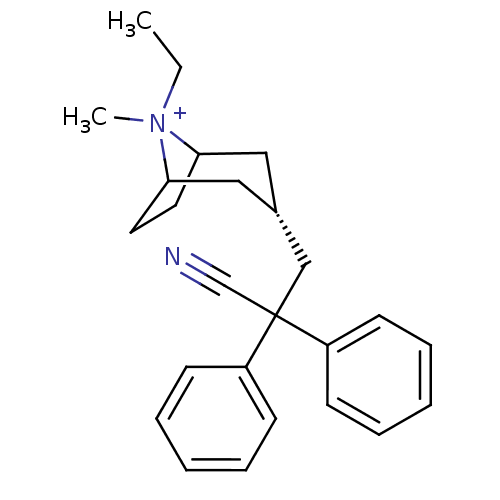
| SMILES | CC[N+]1(C)C2CCC1C[C@@H](CC(C#N)(c1ccccc1)c1ccccc1)C2 |r,wD:9.10,TLB:3:2:9.8.26:6.5,THB:1:2:9.8.26:6.5,10:9:2:6.5,(20.18,4.9,;20.96,3.57,;20.2,2.23,;18.7,2.64,;21.25,1.4,;20.65,.06,;19.47,-.65,;20.46,.69,;22.3,.69,;23.28,-.12,;24.05,-1.45,;25.59,-1.44,;25.99,.05,;26.38,1.55,;26.37,-2.78,;25.6,-4.11,;26.37,-5.44,;27.91,-5.44,;28.68,-4.09,;27.9,-2.76,;27.08,-1.04,;28.17,-2.12,;29.65,-1.71,;30.04,-.22,;28.93,.86,;27.46,.45,;23.01,1.43,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lainé, DI; Yan, H; Xie, H; Davis, RS; Dufour, J; Widdowson, KL; Palovich, MR; Wan, Z; Foley, JJ; Schmidt, DB; Hunsberger, GE; Burman, M; Bacon, AM; Webb, EF; Luttmann, MA; Salmon, M; Sarau, HM; Umbrecht, ST; Landis, PS; Peck, BJ; Busch-Petersen, J Design, synthesis and structure-activity relationship of N-substituted tropane muscarinic acetylcholine receptor antagonists. Bioorg Med Chem Lett22:3366-9 (2012) [PubMed] Article
Lainé, DI; Yan, H; Xie, H; Davis, RS; Dufour, J; Widdowson, KL; Palovich, MR; Wan, Z; Foley, JJ; Schmidt, DB; Hunsberger, GE; Burman, M; Bacon, AM; Webb, EF; Luttmann, MA; Salmon, M; Sarau, HM; Umbrecht, ST; Landis, PS; Peck, BJ; Busch-Petersen, J Design, synthesis and structure-activity relationship of N-substituted tropane muscarinic acetylcholine receptor antagonists. Bioorg Med Chem Lett22:3366-9 (2012) [PubMed] Article