| Citation |  McCoull, W; Addie, MS; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Bowker, SS; Boyd, S; Chapman, S; Davies, RD; Donald, CS; Green, CP; Jenner, C; Kemmitt, PD; Leach, AG; Moody, GC; Gutierrez, PM; Newcombe, NJ; Nowak, T; Packer, MJ; Plowright, AT; Revill, J; Schofield, P; Sheldon, C; Stokes, S; Turnbull, AV; Wang, SJ; Whalley, DP; Wood, JM Identification, optimisation and in vivo evaluation of oxadiazole DGAT-1 inhibitors for the treatment of obesity and diabetes. Bioorg Med Chem Lett22:3873-8 (2012) [PubMed] Article McCoull, W; Addie, MS; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Bowker, SS; Boyd, S; Chapman, S; Davies, RD; Donald, CS; Green, CP; Jenner, C; Kemmitt, PD; Leach, AG; Moody, GC; Gutierrez, PM; Newcombe, NJ; Nowak, T; Packer, MJ; Plowright, AT; Revill, J; Schofield, P; Sheldon, C; Stokes, S; Turnbull, AV; Wang, SJ; Whalley, DP; Wood, JM Identification, optimisation and in vivo evaluation of oxadiazole DGAT-1 inhibitors for the treatment of obesity and diabetes. Bioorg Med Chem Lett22:3873-8 (2012) [PubMed] Article |
|---|
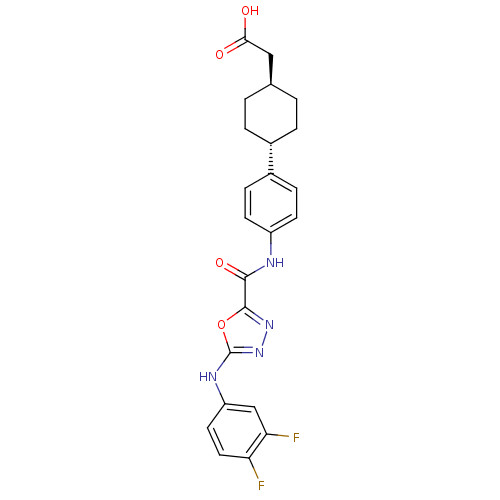
| SMILES | OC(=O)C[C@H]1CC[C@@H](CC1)c1ccc(NC(=O)c2nnc(Nc3ccc(F)c(F)c3)o2)cc1 |r,wU:4.3,wD:7.10,(19.62,-39.47,;18.85,-40.8,;17.31,-40.79,;19.62,-42.14,;21.16,-42.14,;21.93,-40.81,;23.47,-40.82,;24.23,-42.16,;23.46,-43.49,;21.93,-43.48,;25.77,-42.17,;26.55,-40.84,;28.08,-40.85,;28.84,-42.19,;30.38,-42.2,;31.16,-40.87,;30.4,-39.53,;32.71,-40.88,;33.18,-39.41,;34.72,-39.41,;35.19,-40.87,;36.52,-41.64,;37.85,-40.86,;37.84,-39.33,;39.16,-38.55,;40.51,-39.31,;41.84,-38.53,;40.52,-40.85,;41.85,-41.61,;39.19,-41.63,;33.95,-41.78,;28.07,-43.51,;26.54,-43.51,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 McCoull, W; Addie, MS; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Bowker, SS; Boyd, S; Chapman, S; Davies, RD; Donald, CS; Green, CP; Jenner, C; Kemmitt, PD; Leach, AG; Moody, GC; Gutierrez, PM; Newcombe, NJ; Nowak, T; Packer, MJ; Plowright, AT; Revill, J; Schofield, P; Sheldon, C; Stokes, S; Turnbull, AV; Wang, SJ; Whalley, DP; Wood, JM Identification, optimisation and in vivo evaluation of oxadiazole DGAT-1 inhibitors for the treatment of obesity and diabetes. Bioorg Med Chem Lett22:3873-8 (2012) [PubMed] Article
McCoull, W; Addie, MS; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Bowker, SS; Boyd, S; Chapman, S; Davies, RD; Donald, CS; Green, CP; Jenner, C; Kemmitt, PD; Leach, AG; Moody, GC; Gutierrez, PM; Newcombe, NJ; Nowak, T; Packer, MJ; Plowright, AT; Revill, J; Schofield, P; Sheldon, C; Stokes, S; Turnbull, AV; Wang, SJ; Whalley, DP; Wood, JM Identification, optimisation and in vivo evaluation of oxadiazole DGAT-1 inhibitors for the treatment of obesity and diabetes. Bioorg Med Chem Lett22:3873-8 (2012) [PubMed] Article