| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 3A4 |
|---|
| Ligand | BDBM50392452 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_851921 (CHEMBL2156798) |
|---|
| IC50 | 40000±n/a nM |
|---|
| Citation |  Certal, V; Halley, F; Virone-Oddos, A; Thompson, F; Filoche-Rommé, B; El-Ahmad, Y; Carry, JC; Delorme, C; Karlsson, A; Abecassis, PY; Vincent, L; Bonnevaux, H; Nicolas, JP; Morales, R; Michot, N; Vade, I; Louboutin, A; Perron, S; Doerflinger, G; Tric, B; Monget, S; Lengauer, C; Schio, L Preparation and optimization of new 4-(morpholin-4-yl)-(6-oxo-1,6-dihydropyrimidin-2-yl)amide derivatives as PI3Kß inhibitors. Bioorg Med Chem Lett22:6381-4 (2012) [PubMed] Article Certal, V; Halley, F; Virone-Oddos, A; Thompson, F; Filoche-Rommé, B; El-Ahmad, Y; Carry, JC; Delorme, C; Karlsson, A; Abecassis, PY; Vincent, L; Bonnevaux, H; Nicolas, JP; Morales, R; Michot, N; Vade, I; Louboutin, A; Perron, S; Doerflinger, G; Tric, B; Monget, S; Lengauer, C; Schio, L Preparation and optimization of new 4-(morpholin-4-yl)-(6-oxo-1,6-dihydropyrimidin-2-yl)amide derivatives as PI3Kß inhibitors. Bioorg Med Chem Lett22:6381-4 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 3A4 |
|---|
| Name: | Cytochrome P450 3A4 |
|---|
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 57349.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 503 |
|---|
| Sequence: | MALIPDLAMETWLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMF
DMECHKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISI
AEDEEWKRLRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYS
MDVITSTSFGVNIDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICV
FPREVTNFLRKSVKRMKESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSI
IFIFAGYETTSSVLSFIMYELATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVV
NETLRLFPIAMRLERVCKKDVEINGMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFS
KKNKDNIDPYIYTPFGSGPRNCIGMRFALMNMKLALIRVLQNFSFKPCKETQIPLKLSLG
GLLQPEKPVVLKVESRDGTVSGA
|
|
|
|---|
| BDBM50392452 |
|---|
| n/a |
|---|
| Name | BDBM50392452 |
|---|
| Synonyms: | CHEMBL2151922 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H20N4O4 |
|---|
| Mol. Mass. | 344.3651 |
|---|
| SMILES | COc1cccc(NC(=O)Cc2nc(cc(=O)[nH]2)N2CCOCC2)c1 |
|---|
| Structure | 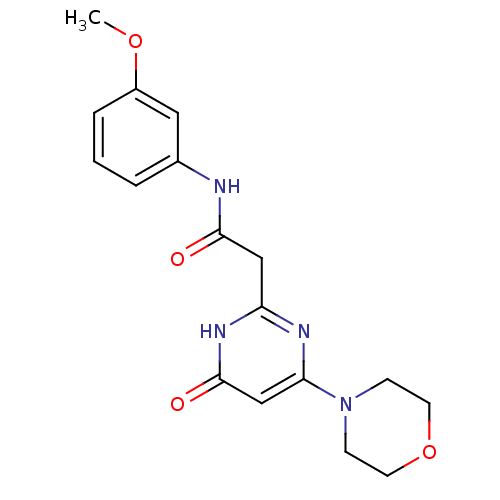
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Certal, V; Halley, F; Virone-Oddos, A; Thompson, F; Filoche-Rommé, B; El-Ahmad, Y; Carry, JC; Delorme, C; Karlsson, A; Abecassis, PY; Vincent, L; Bonnevaux, H; Nicolas, JP; Morales, R; Michot, N; Vade, I; Louboutin, A; Perron, S; Doerflinger, G; Tric, B; Monget, S; Lengauer, C; Schio, L Preparation and optimization of new 4-(morpholin-4-yl)-(6-oxo-1,6-dihydropyrimidin-2-yl)amide derivatives as PI3Kß inhibitors. Bioorg Med Chem Lett22:6381-4 (2012) [PubMed] Article
Certal, V; Halley, F; Virone-Oddos, A; Thompson, F; Filoche-Rommé, B; El-Ahmad, Y; Carry, JC; Delorme, C; Karlsson, A; Abecassis, PY; Vincent, L; Bonnevaux, H; Nicolas, JP; Morales, R; Michot, N; Vade, I; Louboutin, A; Perron, S; Doerflinger, G; Tric, B; Monget, S; Lengauer, C; Schio, L Preparation and optimization of new 4-(morpholin-4-yl)-(6-oxo-1,6-dihydropyrimidin-2-yl)amide derivatives as PI3Kß inhibitors. Bioorg Med Chem Lett22:6381-4 (2012) [PubMed] Article