| Citation |  Barlind, JG; Bauer, UA; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Davies, RD; Eriksson, JW; Hammond, CD; Hovland, R; Johannesson, P; Johansson, MJ; Kemmitt, PD; Lindmark, BT; Morentin Gutierrez, P; Noeske, TA; Nordin, A; O'Donnell, CJ; Petersson, AU; Redzic, A; Turnbull, AV; Vinblad, J Design and optimization of pyrazinecarboxamide-based inhibitors of diacylglycerol acyltransferase 1 (DGAT1) leading to a clinical candidate dimethylpyrazinecarboxamide phenylcyclohexylacetic acid (AZD7687). J Med Chem55:10610-29 (2012) [PubMed] Article Barlind, JG; Bauer, UA; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Davies, RD; Eriksson, JW; Hammond, CD; Hovland, R; Johannesson, P; Johansson, MJ; Kemmitt, PD; Lindmark, BT; Morentin Gutierrez, P; Noeske, TA; Nordin, A; O'Donnell, CJ; Petersson, AU; Redzic, A; Turnbull, AV; Vinblad, J Design and optimization of pyrazinecarboxamide-based inhibitors of diacylglycerol acyltransferase 1 (DGAT1) leading to a clinical candidate dimethylpyrazinecarboxamide phenylcyclohexylacetic acid (AZD7687). J Med Chem55:10610-29 (2012) [PubMed] Article |
|---|
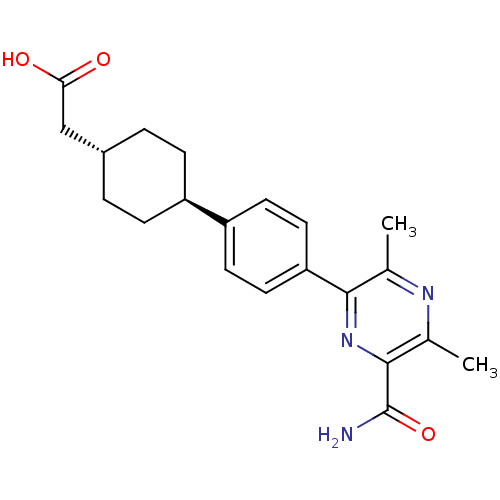
| SMILES | Cc1nc(C)c(nc1C(N)=O)-c1ccc(cc1)[C@H]1CC[C@H](CC(O)=O)CC1 |r,wU:17.18,wD:20.22,(61.46,-10.56,;62.79,-9.79,;64.13,-10.56,;65.46,-9.79,;66.8,-10.56,;65.46,-8.26,;64.13,-7.48,;62.79,-8.25,;61.46,-7.48,;60.12,-8.25,;61.46,-5.94,;66.79,-7.49,;68.12,-8.26,;69.46,-7.49,;69.46,-5.94,;68.12,-5.17,;66.79,-5.94,;70.79,-5.16,;70.78,-3.63,;72.12,-2.86,;73.45,-3.62,;74.79,-2.85,;76.12,-3.62,;77.46,-2.85,;76.12,-5.16,;73.45,-5.17,;72.13,-5.93,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Barlind, JG; Bauer, UA; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Davies, RD; Eriksson, JW; Hammond, CD; Hovland, R; Johannesson, P; Johansson, MJ; Kemmitt, PD; Lindmark, BT; Morentin Gutierrez, P; Noeske, TA; Nordin, A; O'Donnell, CJ; Petersson, AU; Redzic, A; Turnbull, AV; Vinblad, J Design and optimization of pyrazinecarboxamide-based inhibitors of diacylglycerol acyltransferase 1 (DGAT1) leading to a clinical candidate dimethylpyrazinecarboxamide phenylcyclohexylacetic acid (AZD7687). J Med Chem55:10610-29 (2012) [PubMed] Article
Barlind, JG; Bauer, UA; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Davies, RD; Eriksson, JW; Hammond, CD; Hovland, R; Johannesson, P; Johansson, MJ; Kemmitt, PD; Lindmark, BT; Morentin Gutierrez, P; Noeske, TA; Nordin, A; O'Donnell, CJ; Petersson, AU; Redzic, A; Turnbull, AV; Vinblad, J Design and optimization of pyrazinecarboxamide-based inhibitors of diacylglycerol acyltransferase 1 (DGAT1) leading to a clinical candidate dimethylpyrazinecarboxamide phenylcyclohexylacetic acid (AZD7687). J Med Chem55:10610-29 (2012) [PubMed] Article