| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prolyl endopeptidase FAP |
|---|
| Ligand | BDBM50399730 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_876940 (CHEMBL2185388) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Van der Veken, P; Fülöp, V; Rea, D; Gerard, M; Van Elzen, R; Joossens, J; Cheng, JD; Baekelandt, V; De Meester, I; Lambeir, AM; Augustyns, K P2-substituted N-acylprolylpyrrolidine inhibitors of prolyl oligopeptidase: biochemical evaluation, binding mode determination, and assessment in a cellular model of synucleinopathy. J Med Chem55:9856-67 (2012) [PubMed] Article Van der Veken, P; Fülöp, V; Rea, D; Gerard, M; Van Elzen, R; Joossens, J; Cheng, JD; Baekelandt, V; De Meester, I; Lambeir, AM; Augustyns, K P2-substituted N-acylprolylpyrrolidine inhibitors of prolyl oligopeptidase: biochemical evaluation, binding mode determination, and assessment in a cellular model of synucleinopathy. J Med Chem55:9856-67 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prolyl endopeptidase FAP |
|---|
| Name: | Prolyl endopeptidase FAP |
|---|
| Synonyms: | Fap | Fibroblast activation protein (FAP) | SEPR_MOUSE | Seprase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 87949.14 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | P97321 |
|---|
| Residue: | 761 |
|---|
| Sequence: | MKTWLKTVFGVTTLAALALVVICIVLRPSRVYKPEGNTKRALTLKDILNGTFSYKTYFPN
WISEQEYLHQSEDDNIVFYNIETRESYIILSNSTMKSVNATDYGLSPDRQFVYLESDYSK
LWRYSYTATYYIYDLQNGEFVRGYELPRPIQYLCWSPVGSKLAYVYQNNIYLKQRPGDPP
FQITYTGRENRIFNGIPDWVYEEEMLATKYALWWSPDGKFLAYVEFNDSDIPIIAYSYYG
DGQYPRTINIPYPKAGAKNPVVRVFIVDTTYPHHVGPMEVPVPEMIASSDYYFSWLTWVS
SERVCLQWLKRVQNVSVLSICDFREDWHAWECPKNQEHVEESRTGWAGGFFVSTPAFSQD
ATSYYKIFSDKDGYKHIHYIKDTVENAIQITSGKWEAIYIFRVTQDSLFYSSNEFEGYPG
RRNIYRISIGNSPPSKKCVTCHLRKERCQYYTASFSYKAKYYALVCYGPGLPISTLHDGR
TDQEIQVLEENKELENSLRNIQLPKVEIKKLKDGGLTFWYKMILPPQFDRSKKYPLLIQV
YGGPCSQSVKSVFAVNWITYLASKEGIVIALVDGRGTAFQGDKFLHAVYRKLGVYEVEDQ
LTAVRKFIEMGFIDEERIAIWGWSYGGYVSSLALASGTGLFKCGIAVAPVSSWEYYASIY
SERFMGLPTKDDNLEHYKNSTVMARAEYFRNVDYLLIHGTADDNVHFQNSAQIAKALVNA
QVDFQAMWYSDQNHGISSGRSQNHLYTHMTHFLKQCFSLSD
|
|
|
|---|
| BDBM50399730 |
|---|
| n/a |
|---|
| Name | BDBM50399730 |
|---|
| Synonyms: | CHEMBL2178967 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H30BrN5O2 |
|---|
| Mol. Mass. | 536.463 |
|---|
| SMILES | Brc1ccc(cc1)-c1cn(nn1)[C@H]1C[C@H](N(C1)C(=O)CCCc1ccccc1)C(=O)N1CCCC1 |r| |
|---|
| Structure | 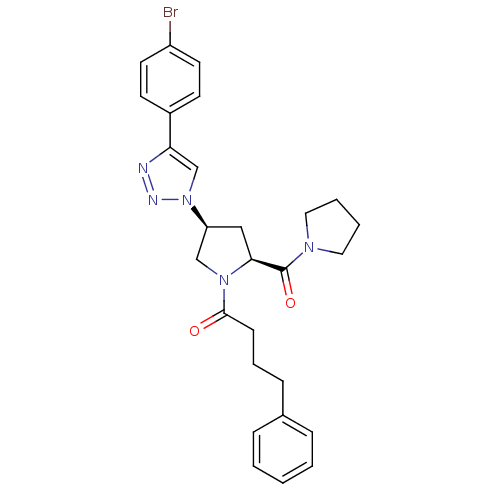
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Van der Veken, P; Fülöp, V; Rea, D; Gerard, M; Van Elzen, R; Joossens, J; Cheng, JD; Baekelandt, V; De Meester, I; Lambeir, AM; Augustyns, K P2-substituted N-acylprolylpyrrolidine inhibitors of prolyl oligopeptidase: biochemical evaluation, binding mode determination, and assessment in a cellular model of synucleinopathy. J Med Chem55:9856-67 (2012) [PubMed] Article
Van der Veken, P; Fülöp, V; Rea, D; Gerard, M; Van Elzen, R; Joossens, J; Cheng, JD; Baekelandt, V; De Meester, I; Lambeir, AM; Augustyns, K P2-substituted N-acylprolylpyrrolidine inhibitors of prolyl oligopeptidase: biochemical evaluation, binding mode determination, and assessment in a cellular model of synucleinopathy. J Med Chem55:9856-67 (2012) [PubMed] Article