

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Ligand | BDBM50417100 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_665340 (CHEMBL1261017) | ||
| Ki | 43.65±n/a nM | ||
| Citation |  de Kloe, GE; Retra, K; Geitmann, M; Källblad, P; Nahar, T; van Elk, R; Smit, AB; van Muijlwijk-Koezen, JE; Leurs, R; Irth, H; Danielson, UH; de Esch, IJ Surface plasmon resonance biosensor based fragment screening using acetylcholine binding protein identifies ligand efficiency hot spots (LE hot spots) by deconstruction of nicotinic acetylcholine receptora7 ligands. J Med Chem53:7192-201 (2010) [PubMed] Article de Kloe, GE; Retra, K; Geitmann, M; Källblad, P; Nahar, T; van Elk, R; Smit, AB; van Muijlwijk-Koezen, JE; Leurs, R; Irth, H; Danielson, UH; de Esch, IJ Surface plasmon resonance biosensor based fragment screening using acetylcholine binding protein identifies ligand efficiency hot spots (LE hot spots) by deconstruction of nicotinic acetylcholine receptora7 ligands. J Med Chem53:7192-201 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-7 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Synonyms: | ACHA7_HUMAN | CHRNA7 | Cholinergic, Nicotinic Alpha7 | NACHRA7 | Neuronal acetylcholine receptor protein alpha-7 subunit | Neuronal acetylcholine receptor subunit alpha-7 (nAChR-alpha 7) | Nicotinic acetylcholine receptor alpha-7 (alpha7 nAChR) | ||
| Type: | n/a | ||
| Mol. Mass.: | 56448.33 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | CHRNA7 (NACHRA7) | ||
| Residue: | 502 | ||
| Sequence: |
| ||
| BDBM50417100 | |||
| n/a | |||
| Name | BDBM50417100 | ||
| Synonyms: | CHEMBL1258125 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H21N3OS | ||
| Mol. Mass. | 327.444 | ||
| SMILES | O=C(N[C@@H]1C2CCN(CC2)[C@H]1Cc1cccnc1)c1cccs1 |r,wU:3.2,wD:10.12,TLB:11:10:6.5:8.9,THB:2:3:6.5:8.9,(18.11,-17.12,;18.13,-15.58,;16.81,-14.79,;15.46,-15.54,;14,-14.9,;12.64,-15.51,;12.37,-16.9,;13.74,-16.27,;13.48,-14.37,;13.93,-13.26,;15.27,-16.92,;16.03,-18.27,;15.25,-19.59,;13.71,-19.57,;12.93,-20.9,;13.69,-22.24,;15.24,-22.25,;16.01,-20.92,;19.48,-14.83,;19.65,-13.3,;21.16,-13,;21.91,-14.34,;20.87,-15.48,)| | ||
| Structure | 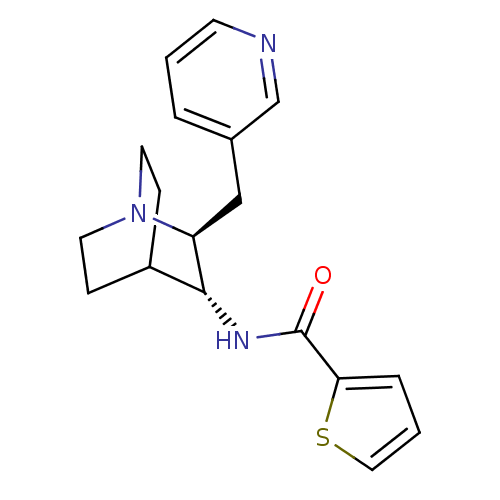
| ||