| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Muscarinic acetylcholine receptor M3 |
|---|
| Ligand | BDBM50194640 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_936428 (CHEMBL2317637) |
|---|
| Ki | 0.300000±n/a nM |
|---|
| Citation |  Zheng, G; Smith, AM; Huang, X; Subramanian, KL; Siripurapu, KB; Deaciuc, A; Zhan, CG; Dwoskin, LP Structural modifications to tetrahydropyridine-3-carboxylate esters en route to the discovery of M5-preferring muscarinic receptor orthosteric antagonists. J Med Chem56:1693-703 (2013) [PubMed] Article Zheng, G; Smith, AM; Huang, X; Subramanian, KL; Siripurapu, KB; Deaciuc, A; Zhan, CG; Dwoskin, LP Structural modifications to tetrahydropyridine-3-carboxylate esters en route to the discovery of M5-preferring muscarinic receptor orthosteric antagonists. J Med Chem56:1693-703 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Muscarinic acetylcholine receptor M3 |
|---|
| Name: | Muscarinic acetylcholine receptor M3 |
|---|
| Synonyms: | ACM3_HUMAN | CHRM3 | Cholinergic, muscarinic M3 | Muscarinic Receptors M3 | Muscarinic receptor M3 | RecName: Full=Muscarinic acetylcholine receptor M3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 66151.03 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P20309 |
|---|
| Residue: | 590 |
|---|
| Sequence: | MTLHNNSTTSPLFPNISSSWIHSPSDAGLPPGTVTHFGSYNVSRAAGNFSSPDGTTDDPL
GGHTVWQVVFIAFLTGILALVTIIGNILVIVSFKVNKQLKTVNNYFLLSLACADLIIGVI
SMNLFTTYIIMNRWALGNLACDLWLAIDYVASNASVMNLLVISFDRYFSITRPLTYRAKR
TTKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRTVPPGECFIQFLSEPTITFGTAIAAF
YMPVTIMTILYWRIYKETEKRTKELAGLQASGTEAETENFVHPTGSSRSCSSYELQQQSM
KRSNRRKYGRCHFWFTTKSWKPSSEQMDQDHSSSDSWNNNDAAASLENSASSDEEDIGSE
TRAIYSIVLKLPGHSTILNSTKLPSSDNLQVPEEELGMVDLERKADKLQAQKSVDDGGSF
PKSFSKLPIQLESAVDTAKTSDVNSSVGKSTATLPLSFKEATLAKRFALKTRSQITKRKR
MSLVKEKKAAQTLSAILLAFIITWTPYNIMVLVNTFCDSCIPKTFWNLGYWLCYINSTVN
PVCYALCNKTFRTTFKMLLLCQCDKKKRRKQQYQQRQSVIFHKRAPEQAL
|
|
|
|---|
| BDBM50194640 |
|---|
| n/a |
|---|
| Name | BDBM50194640 |
|---|
| Synonyms: | (2R)-1-((2S,4R)-4-hydroxy-1-[3,3,3-tris(4-fluorophenyl)propanoyl]pyrrolidine-2-yl)carbonyl-N-(4-piperidinylmethyl)pyrrolidine-2-carboxamide | CHEMBL215180 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C37H41F3N4O4 |
|---|
| Mol. Mass. | 662.741 |
|---|
| SMILES | O[C@@H]1C[C@H](N(C1)C(=O)CC(c1ccc(F)cc1)(c1ccc(F)cc1)c1ccc(F)cc1)C(=O)N1CCC[C@@H]1C(=O)NCC1CCNCC1 |
|---|
| Structure | 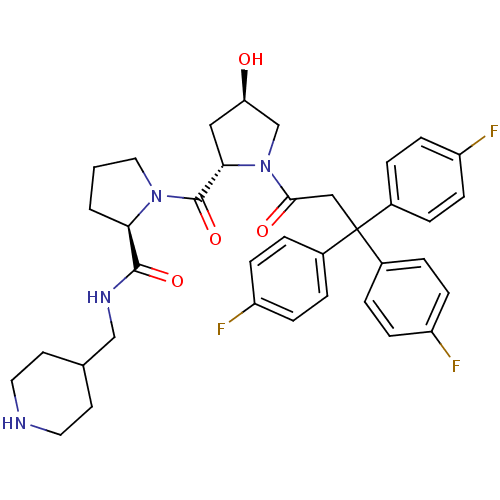
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zheng, G; Smith, AM; Huang, X; Subramanian, KL; Siripurapu, KB; Deaciuc, A; Zhan, CG; Dwoskin, LP Structural modifications to tetrahydropyridine-3-carboxylate esters en route to the discovery of M5-preferring muscarinic receptor orthosteric antagonists. J Med Chem56:1693-703 (2013) [PubMed] Article
Zheng, G; Smith, AM; Huang, X; Subramanian, KL; Siripurapu, KB; Deaciuc, A; Zhan, CG; Dwoskin, LP Structural modifications to tetrahydropyridine-3-carboxylate esters en route to the discovery of M5-preferring muscarinic receptor orthosteric antagonists. J Med Chem56:1693-703 (2013) [PubMed] Article