| Citation |  Décor, A; Grand-Maître, C; Hucke, O; O'Meara, J; Kuhn, C; Constantineau-Forget, L; Brochu, C; Malenfant, E; Bertrand-Laperle, M; Bordeleau, J; Ghiro, E; Pesant, M; Fazal, G; Gorys, V; Little, M; Boucher, C; Bordeleau, S; Turcotte, P; Guo, T; Garneau, M; Spickler, C; Gauthier, A Design, synthesis and biological evaluation of novel aminothiazoles as antiviral compounds acting against human rhinovirus. Bioorg Med Chem Lett23:3841-7 (2013) [PubMed] Article Décor, A; Grand-Maître, C; Hucke, O; O'Meara, J; Kuhn, C; Constantineau-Forget, L; Brochu, C; Malenfant, E; Bertrand-Laperle, M; Bordeleau, J; Ghiro, E; Pesant, M; Fazal, G; Gorys, V; Little, M; Boucher, C; Bordeleau, S; Turcotte, P; Guo, T; Garneau, M; Spickler, C; Gauthier, A Design, synthesis and biological evaluation of novel aminothiazoles as antiviral compounds acting against human rhinovirus. Bioorg Med Chem Lett23:3841-7 (2013) [PubMed] Article |
|---|
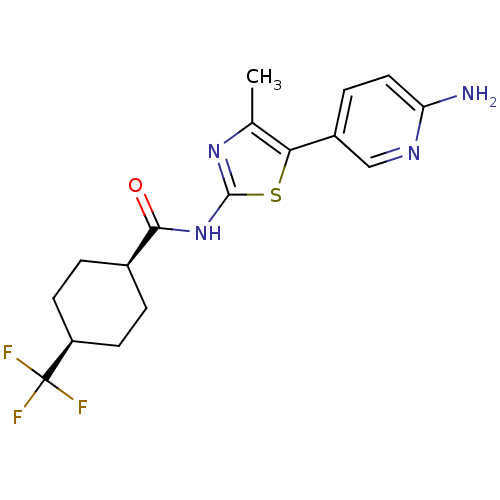
| SMILES | Cc1nc(NC(=O)[C@H]2CC[C@H](CC2)C(F)(F)F)sc1-c1ccc(N)nc1 |r,wU:7.6,10.13,(28.23,-22.96,;29.38,-23.98,;30.89,-23.66,;31.66,-24.99,;33.2,-25.14,;33.83,-26.55,;32.93,-27.8,;35.36,-26.7,;36.25,-25.45,;37.79,-25.61,;38.42,-27.02,;37.51,-28.27,;35.99,-28.11,;39.95,-27.18,;40.58,-28.59,;40.86,-25.94,;41.49,-27.18,;30.64,-26.14,;29.23,-25.51,;27.89,-26.28,;26.56,-25.52,;25.23,-26.29,;25.23,-27.83,;23.89,-28.6,;26.56,-28.6,;27.9,-27.83,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Décor, A; Grand-Maître, C; Hucke, O; O'Meara, J; Kuhn, C; Constantineau-Forget, L; Brochu, C; Malenfant, E; Bertrand-Laperle, M; Bordeleau, J; Ghiro, E; Pesant, M; Fazal, G; Gorys, V; Little, M; Boucher, C; Bordeleau, S; Turcotte, P; Guo, T; Garneau, M; Spickler, C; Gauthier, A Design, synthesis and biological evaluation of novel aminothiazoles as antiviral compounds acting against human rhinovirus. Bioorg Med Chem Lett23:3841-7 (2013) [PubMed] Article
Décor, A; Grand-Maître, C; Hucke, O; O'Meara, J; Kuhn, C; Constantineau-Forget, L; Brochu, C; Malenfant, E; Bertrand-Laperle, M; Bordeleau, J; Ghiro, E; Pesant, M; Fazal, G; Gorys, V; Little, M; Boucher, C; Bordeleau, S; Turcotte, P; Guo, T; Garneau, M; Spickler, C; Gauthier, A Design, synthesis and biological evaluation of novel aminothiazoles as antiviral compounds acting against human rhinovirus. Bioorg Med Chem Lett23:3841-7 (2013) [PubMed] Article