

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | P2X purinoceptor 7 | ||
| Ligand | BDBM50437329 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_972155 (CHEMBL2411084) | ||
| IC50 | 7700±n/a nM | ||
| Citation |  Battilocchio, C; Guetzoyan, L; Cervetto, C; Di Cesare Mannelli, L; Frattaroli, D; Baxendale, IR; Maura, G; Rossi, A; Sautebin, L; Biava, M; Ghelardini, C; Marcoli, M; Ley, SV Flow Synthesis and Biological Studies of an Analgesic Adamantane Derivative That Inhibits P2X7-Evoked Glutamate Release. ACS Med Chem Lett4:704-9 (2013) [PubMed] Article Battilocchio, C; Guetzoyan, L; Cervetto, C; Di Cesare Mannelli, L; Frattaroli, D; Baxendale, IR; Maura, G; Rossi, A; Sautebin, L; Biava, M; Ghelardini, C; Marcoli, M; Ley, SV Flow Synthesis and Biological Studies of an Analgesic Adamantane Derivative That Inhibits P2X7-Evoked Glutamate Release. ACS Med Chem Lett4:704-9 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| P2X purinoceptor 7 | |||
| Name: | P2X purinoceptor 7 | ||
| Synonyms: | ATP receptor | P2RX7_RAT | P2X purinoceptor 7 | P2X purinoceptor 7 (P2RX7) | P2X purinoceptor 7 (P2X7) | P2X7 | P2X7 rat | P2Z receptor | P2rx7 | Purinergic receptor | ||
| Type: | Protein | ||
| Mol. Mass.: | 68403.50 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | Q64663 | ||
| Residue: | 595 | ||
| Sequence: |
| ||
| BDBM50437329 | |||
| n/a | |||
| Name | BDBM50437329 | ||
| Synonyms: | CHEMBL2407630 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H26N2O2 | ||
| Mol. Mass. | 338.4433 | ||
| SMILES | O=C(NC1(C2CC3CC(C2)CC1C3)C(=O)NC1CC1)c1ccccc1 |TLB:2:3:5.6.12:9.8.10,2:3:5:9.8.7,13:3:5:9.8.7,THB:13:3:5.6.12:9.8.10,7:6:3:9.8.10,7:8:3:5.6.12,10:11:5:9.8.7,10:8:5:3.12.11,(22.9,-26.25,;22.14,-24.91,;20.6,-24.91,;19.84,-23.58,;18.44,-23.02,;16.93,-23.44,;18.13,-22.16,;18.12,-20.68,;19.47,-20.2,;18.43,-21.43,;20.87,-20.78,;20.86,-22.31,;19.46,-22.65,;19.06,-24.9,;19.83,-26.24,;17.52,-24.9,;16.75,-26.23,;15.41,-27.01,;16.74,-27.78,;22.91,-23.58,;24.45,-23.6,;25.23,-22.27,;24.46,-20.93,;22.91,-20.93,;22.14,-22.26,)| | ||
| Structure | 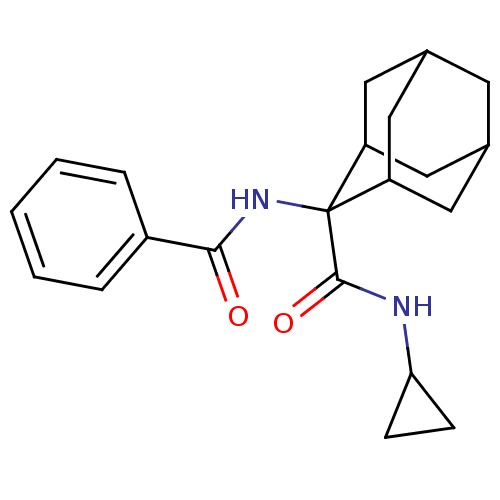
| ||