| Citation |  He, S; Hong, Q; Lai, Z; Wu, Z; Yu, Y; Kim, DW; Ting, PC; Kuethe, JT; Yang, GX; Jian, T; Liu, J; Guiadeen, D; Krikorian, AD; Sperbeck, DM; Sonatore, LM; Wiltsie, J; Chung, CC; Gibson, JT; Lisnock, J; Murphy, BA; Gorski, JN; Liu, J; Chen, D; Chen, X; Wolff, M; Tong, SX; Madeira, M; Karanam, BV; Shen, DM; Balkovec, JM; Pinto, S; Nargund, RP; DeVita, RJ Potent DGAT1 Inhibitors in the Benzimidazole Class with a Pyridyl-oxy-cyclohexanecarboxylic Acid Moiety. ACS Med Chem Lett4:773-8 (2013) [PubMed] Article He, S; Hong, Q; Lai, Z; Wu, Z; Yu, Y; Kim, DW; Ting, PC; Kuethe, JT; Yang, GX; Jian, T; Liu, J; Guiadeen, D; Krikorian, AD; Sperbeck, DM; Sonatore, LM; Wiltsie, J; Chung, CC; Gibson, JT; Lisnock, J; Murphy, BA; Gorski, JN; Liu, J; Chen, D; Chen, X; Wolff, M; Tong, SX; Madeira, M; Karanam, BV; Shen, DM; Balkovec, JM; Pinto, S; Nargund, RP; DeVita, RJ Potent DGAT1 Inhibitors in the Benzimidazole Class with a Pyridyl-oxy-cyclohexanecarboxylic Acid Moiety. ACS Med Chem Lett4:773-8 (2013) [PubMed] Article |
|---|
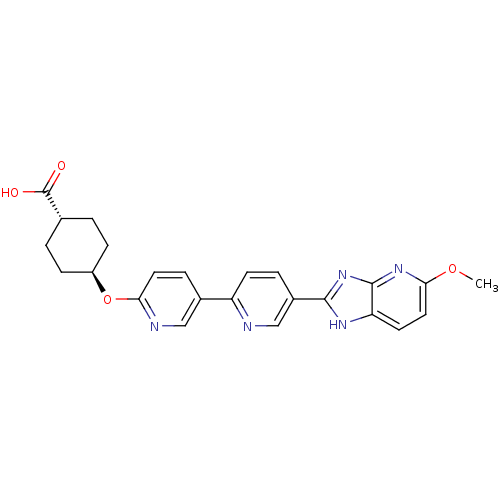
| SMILES | COc1ccc2[nH]c(nc2n1)-c1ccc(nc1)-c1ccc(O[C@H]2CC[C@@H](CC2)C(O)=O)nc1 |r,wU:22.24,wD:25.31,(24.23,-45.19,;24.23,-46.73,;25.57,-47.5,;25.57,-49.04,;26.89,-49.8,;28.22,-49.04,;29.69,-49.52,;30.6,-48.27,;29.69,-47.02,;28.22,-47.5,;26.89,-46.72,;32.14,-48.27,;32.9,-46.94,;34.44,-46.93,;35.21,-48.27,;34.44,-49.61,;32.9,-49.61,;36.75,-48.27,;37.51,-46.94,;39.05,-46.93,;39.83,-48.27,;41.37,-48.27,;42.14,-49.6,;43.67,-49.58,;44.44,-50.92,;43.67,-52.25,;42.13,-52.25,;41.37,-50.92,;44.45,-53.6,;43.68,-54.93,;45.99,-53.6,;39.06,-49.61,;37.52,-49.61,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 He, S; Hong, Q; Lai, Z; Wu, Z; Yu, Y; Kim, DW; Ting, PC; Kuethe, JT; Yang, GX; Jian, T; Liu, J; Guiadeen, D; Krikorian, AD; Sperbeck, DM; Sonatore, LM; Wiltsie, J; Chung, CC; Gibson, JT; Lisnock, J; Murphy, BA; Gorski, JN; Liu, J; Chen, D; Chen, X; Wolff, M; Tong, SX; Madeira, M; Karanam, BV; Shen, DM; Balkovec, JM; Pinto, S; Nargund, RP; DeVita, RJ Potent DGAT1 Inhibitors in the Benzimidazole Class with a Pyridyl-oxy-cyclohexanecarboxylic Acid Moiety. ACS Med Chem Lett4:773-8 (2013) [PubMed] Article
He, S; Hong, Q; Lai, Z; Wu, Z; Yu, Y; Kim, DW; Ting, PC; Kuethe, JT; Yang, GX; Jian, T; Liu, J; Guiadeen, D; Krikorian, AD; Sperbeck, DM; Sonatore, LM; Wiltsie, J; Chung, CC; Gibson, JT; Lisnock, J; Murphy, BA; Gorski, JN; Liu, J; Chen, D; Chen, X; Wolff, M; Tong, SX; Madeira, M; Karanam, BV; Shen, DM; Balkovec, JM; Pinto, S; Nargund, RP; DeVita, RJ Potent DGAT1 Inhibitors in the Benzimidazole Class with a Pyridyl-oxy-cyclohexanecarboxylic Acid Moiety. ACS Med Chem Lett4:773-8 (2013) [PubMed] Article