| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cyclin-dependent kinase 2/G1/S-specific cyclin-E1 |
|---|
| Ligand | BDBM6860 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1289052 (CHEMBL3118310) |
|---|
| IC50 | 520±n/a nM |
|---|
| Citation |  Reddy, MV; Akula, B; Cosenza, SC; Athuluridivakar, S; Mallireddigari, MR; Pallela, VR; Billa, VK; Subbaiah, DR; Bharathi, EV; Vasquez-Del Carpio, R; Padgaonkar, A; Baker, SJ; Reddy, EP Discovery of 8-cyclopentyl-2-[4-(4-methyl-piperazin-1-yl)-phenylamino]-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidine-6-carbonitrile (7x) as a potent inhibitor of cyclin-dependent kinase 4 (CDK4) and AMPK-related kinase 5 (ARK5). J Med Chem57:578-99 (2014) [PubMed] Article Reddy, MV; Akula, B; Cosenza, SC; Athuluridivakar, S; Mallireddigari, MR; Pallela, VR; Billa, VK; Subbaiah, DR; Bharathi, EV; Vasquez-Del Carpio, R; Padgaonkar, A; Baker, SJ; Reddy, EP Discovery of 8-cyclopentyl-2-[4-(4-methyl-piperazin-1-yl)-phenylamino]-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidine-6-carbonitrile (7x) as a potent inhibitor of cyclin-dependent kinase 4 (CDK4) and AMPK-related kinase 5 (ARK5). J Med Chem57:578-99 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cyclin-dependent kinase 2/G1/S-specific cyclin-E1 |
|---|
| Name: | Cyclin-dependent kinase 2/G1/S-specific cyclin-E1 |
|---|
| Synonyms: | CDK2/CycE | CDK2/Cyclin E | CDK2/E | CDK2/E1 | Cyclin-Dependent Kinase 2 (CDK2) | Cyclin-dependent kinase 2/G1/S-specific cyclin E1 | Cyclin-dependent kinase 2/cyclin E1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Cyclin-dependent kinase 2 |
|---|
| Synonyms: | CDK2 | CDK2-Kinase | CDK2_HUMAN | CDKN2 | Cell division protein kinase 2 | Cyclin-dependent kinase 2 (CDK2) | Protein cereblon/Cyclin-dependent kinase 2 | p33 protein kinase |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 33938.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P24941 |
|---|
| Residue: | 298 |
|---|
| Sequence: | MENFQKVEKIGEGTYGVVYKARNKLTGEVVALKKIRLDTETEGVPSTAIREISLLKELNH
PNIVKLLDVIHTENKLYLVFEFLHQDLKKFMDASALTGIPLPLIKSYLFQLLQGLAFCHS
HRVLHRDLKPQNLLINTEGAIKLADFGLARAFGVPVRTYTHEVVTLWYRAPEILLGCKYY
STAVDIWSLGCIFAEMVTRRALFPGDSEIDQLFRIFRTLGTPDEVVWPGVTSMPDYKPSF
PKWARQDFSKVVPPLDEDGRSLLSQMLHYDPNKRISAKAALAHPFFQDVTKPVPHLRL
|
|
|
|---|
| Component 2 |
| Name: | G1/S-specific cyclin-E1 |
|---|
| Synonyms: | CCNE | CCNE1 | CCNE1_HUMAN |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 47073.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 410 |
|---|
| Sequence: | MPRERRERDAKERDTMKEDGGAEFSARSRKRKANVTVFLQDPDEEMAKIDRTARDQCGSQ
PWDNNAVCADPCSLIPTPDKEDDDRVYPNSTCKPRIIAPSRGSPLPVLSWANREEVWKIM
LNKEKTYLRDQHFLEQHPLLQPKMRAILLDWLMEVCEVYKLHRETFYLAQDFFDRYMATQ
ENVVKTLLQLIGISSLFIAAKLEEIYPPKLHQFAYVTDGACSGDEILTMELMIMKALKWR
LSPLTIVSWLNVYMQVAYLNDLHEVLLPQYPQQIFIQIAELLDLCVLDVDCLEFPYGILA
ASALYHFSSSELMQKVSGYQWCDIENCVKWMVPFAMVIRETGSSKLKHFRGVADEDAHNI
QTHRDSLDLLDKARAKKAMLSEQNRASPLPSGLLTPPQSGKKQSSGPEMA
|
|
|
|---|
| BDBM6860 |
|---|
| n/a |
|---|
| Name | BDBM6860 |
|---|
| Synonyms: | 6-bromo-3,13,23-triazahexacyclo[14.7.0.0^{2,10}.0^{4,9}.0^{11,15}.0^{17,22}]tricosa-1(16),2(10),4(9),5,7,11(15),17(22),18,20-nonaene-12,14-dione | Indolocarbazole deriv. 4(d) |
|---|
| Type | Small Organic Molecule |
|---|
| Emp. Form. | C20H10BrN3O2 |
|---|
| Mol. Mass. | 404.216 |
|---|
| SMILES | Brc1ccc2c(c1)[nH]c1c3[nH]c4ccccc4c3c3C(=O)NC(=O)c3c21 |
|---|
| Structure | 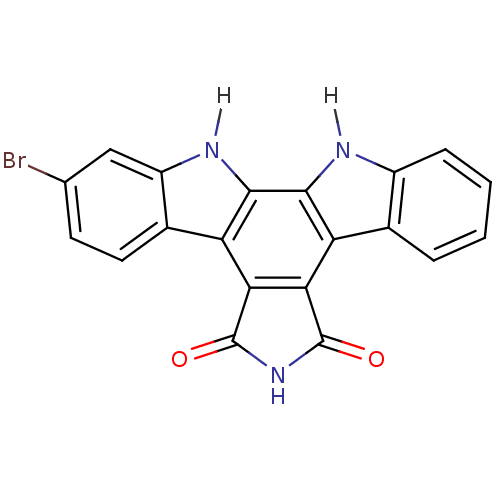
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Reddy, MV; Akula, B; Cosenza, SC; Athuluridivakar, S; Mallireddigari, MR; Pallela, VR; Billa, VK; Subbaiah, DR; Bharathi, EV; Vasquez-Del Carpio, R; Padgaonkar, A; Baker, SJ; Reddy, EP Discovery of 8-cyclopentyl-2-[4-(4-methyl-piperazin-1-yl)-phenylamino]-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidine-6-carbonitrile (7x) as a potent inhibitor of cyclin-dependent kinase 4 (CDK4) and AMPK-related kinase 5 (ARK5). J Med Chem57:578-99 (2014) [PubMed] Article
Reddy, MV; Akula, B; Cosenza, SC; Athuluridivakar, S; Mallireddigari, MR; Pallela, VR; Billa, VK; Subbaiah, DR; Bharathi, EV; Vasquez-Del Carpio, R; Padgaonkar, A; Baker, SJ; Reddy, EP Discovery of 8-cyclopentyl-2-[4-(4-methyl-piperazin-1-yl)-phenylamino]-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidine-6-carbonitrile (7x) as a potent inhibitor of cyclin-dependent kinase 4 (CDK4) and AMPK-related kinase 5 (ARK5). J Med Chem57:578-99 (2014) [PubMed] Article