| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sucrase-isomaltase, intestinal |
|---|
| Ligand | BDBM50028181 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1435477 (CHEMBL3384692) |
|---|
| IC50 | 1500±n/a nM |
|---|
| Citation |  Ghisaidoobe, AT; van den Berg, RJ; Butt, SS; Strijland, A; Donker-Koopman, WE; Scheij, S; van den Nieuwendijk, AM; Koomen, GJ; van Loevezijn, A; Leemhuis, M; Wennekes, T; van der Stelt, M; van der Marel, GA; van Boeckel, CA; Aerts, JM; Overkleeft, HS Identification and development of biphenyl substituted iminosugars as improved dual glucosylceramide synthase/neutral glucosylceramidase inhibitors. J Med Chem57:9096-104 (2014) [PubMed] Article Ghisaidoobe, AT; van den Berg, RJ; Butt, SS; Strijland, A; Donker-Koopman, WE; Scheij, S; van den Nieuwendijk, AM; Koomen, GJ; van Loevezijn, A; Leemhuis, M; Wennekes, T; van der Stelt, M; van der Marel, GA; van Boeckel, CA; Aerts, JM; Overkleeft, HS Identification and development of biphenyl substituted iminosugars as improved dual glucosylceramide synthase/neutral glucosylceramidase inhibitors. J Med Chem57:9096-104 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Sucrase-isomaltase, intestinal |
|---|
| Name: | Sucrase-isomaltase, intestinal |
|---|
| Synonyms: | Alpha glucosidase | Isomaltase | SI | SUIS_HUMAN | Sucrase | Sucrase-isomaltase | Sucrase-isomaltase, intestinal |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 209423.23 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1435477 |
|---|
| Residue: | 1827 |
|---|
| Sequence: | MARKKFSGLEISLIVLFVIVTIIAIALIVVLATKTPAVDEISDSTSTPATTRVTTNPSDS
GKCPNVLNDPVNVRINCIPEQFPTEGICAQRGCCWRPWNDSLIPWCFFVDNHGYNVQDMT
TTSIGVEAKLNRIPSPTLFGNDINSVLFTTQNQTPNRFRFKITDPNNRRYEVPHQYVKEF
TGPTVSDTLYDVKVAQNPFSIQVIRKSNGKTLFDTSIGPLVYSDQYLQISTRLPSDYIYG
IGEQVHKRFRHDLSWKTWPIFTRDQLPGDNNNNLYGHQTFFMCIEDTSGKSFGVFLMNSN
AMEIFIQPTPIVTYRVTGGILDFYILLGDTPEQVVQQYQQLVGLPAMPAYWNLGFQLSRW
NYKSLDVVKEVVRRNREAGIPFDTQVTDIDYMEDKKDFTYDQVAFNGLPQFVQDLHDHGQ
KYVIILDPAISIGRRANGTTYATYERGNTQHVWINESDGSTPIIGEVWPGLTVYPDFTNP
NCIDWWANECSIFHQEVQYDGLWIDMNEVSSFIQGSTKGCNVNKLNYPPFTPDILDKLMY
SKTICMDAVQNWGKQYDVHSLYGYSMAIATEQAVQKVFPNKRSFILTRSTFAGSGRHAAH
WLGDNTASWEQMEWSITGMLEFSLFGIPLVGADICGFVAETTEELCRRWMQLGAFYPFSR
NHNSDGYEHQDPAFFGQNSLLVKSSRQYLTIRYTLLPFLYTLFYKAHVFGETVARPVLHE
FYEDTNSWIEDTEFLWGPALLITPVLKQGADTVSAYIPDAIWYDYESGAKRPWRKQRVDM
YLPADKIGLHLRGGYIIPIQEPDVTTTASRKNPLGLIVALGENNTAKGDFFWDDGETKDT
IQNGNYILYTFSVSNNTLDIVCTHSSYQEGTTLAFQTVKILGLTDSVTEVRVAENNQPMN
AHSNFTYDASNQVLLIADLKLNLGRNFSVQWNQIFSENERFNCYPDADLATEQKCTQRGC
VWRTGSSLSKAPECYFPRQDNSYSVNSARYSSMGITADLQLNTANARIKLPSDPISTLRV
EVKYHKNDMLQFKIYDPQKKRYEVPVPLNIPTTPISTYEDRLYDVEIKENPFGIQIRRRS
SGRVIWDSWLPGFAFNDQFIQISTRLPSEYIYGFGEVEHTAFKRDLNWNTWGMFTRDQPP
GYKLNSYGFHPYYMALEEEGNAHGVFLLNSNAMDVTFQPTPALTYRTVGGILDFYMFLGP
TPEVATKQYHEVIGHPVMPAYWALGFQLCRYGYANTSEVRELYDAMVAANIPYDVQYTDI
DYMERQLDFTIGEAFQDLPQFVDKIRGEGMRYIIILDPAISGNETKTYPAFERGQQNDVF
VKWPNTNDICWAKVWPDLPNITIDKTLTEDEAVNASRAHVAFPDFFRTSTAEWWAREIVD
FYNEKMKFDGLWIDMNEPSSFVNGTTTNQCRNDELNYPPYFPELTKRTDGLHFRTICMEA
EQILSDGTSVLHYDVHNLYGWSQMKPTHDALQKTTGKRGIVISRSTYPTSGRWGGHWLGD
NYARWDNMDKSIIGMMEFSLFGMSYTGADICGFFNNSEYHLCTRWMQLGAFYPYSRNHNI
ANTRRQDPASWNETFAEMSRNILNIRYTLLPYFYTQMHEIHANGGTVIRPLLHEFFDEKP
TWDIFKQFLWGPAFMVTPVLEPYVQTVNAYVPNARWFDYHTGKDIGVRGQFQTFNASYDT
INLHVRGGHILPCQEPAQNTFYSRQKHMKLIVAADDNQMAQGSLFWDDGESIDTYERDLY
LSVQFNLNQTTLTSTILKRGYINKSETRLGSLHVWGKGTTPVNAVTLTYNGNKNSLPFNE
DTTNMILRIDLTTHNVTLEEPIEINWS
|
|
|
|---|
| BDBM50028181 |
|---|
| n/a |
|---|
| Name | BDBM50028181 |
|---|
| Synonyms: | CHEMBL3354626 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H32F3NO5 |
|---|
| Mol. Mass. | 483.5205 |
|---|
| SMILES | OC[C@@H]1[C@@H](O)[C@H](O)[C@@H](O)CN1CCCCCOCc1ccc(cc1C(F)(F)F)-c1ccccc1 |r| |
|---|
| Structure | 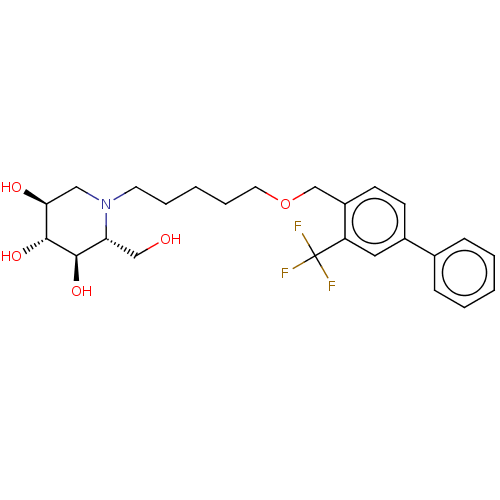
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ghisaidoobe, AT; van den Berg, RJ; Butt, SS; Strijland, A; Donker-Koopman, WE; Scheij, S; van den Nieuwendijk, AM; Koomen, GJ; van Loevezijn, A; Leemhuis, M; Wennekes, T; van der Stelt, M; van der Marel, GA; van Boeckel, CA; Aerts, JM; Overkleeft, HS Identification and development of biphenyl substituted iminosugars as improved dual glucosylceramide synthase/neutral glucosylceramidase inhibitors. J Med Chem57:9096-104 (2014) [PubMed] Article
Ghisaidoobe, AT; van den Berg, RJ; Butt, SS; Strijland, A; Donker-Koopman, WE; Scheij, S; van den Nieuwendijk, AM; Koomen, GJ; van Loevezijn, A; Leemhuis, M; Wennekes, T; van der Stelt, M; van der Marel, GA; van Boeckel, CA; Aerts, JM; Overkleeft, HS Identification and development of biphenyl substituted iminosugars as improved dual glucosylceramide synthase/neutral glucosylceramidase inhibitors. J Med Chem57:9096-104 (2014) [PubMed] Article