| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 1/3/4/6/7 |
|---|
| Ligand | BDBM50047416 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1450204 (CHEMBL3375767) |
|---|
| IC50 | 3.6±n/a nM |
|---|
| Citation |  McCoull, W; Barton, P; Brown, AJ; Bowker, SS; Cameron, J; Clarke, DS; Davies, RD; Dossetter, AG; Ertan, A; Fenwick, M; Green, C; Holmes, JL; Martin, N; Masters, D; Moore, JE; Newcombe, NJ; Newton, C; Pointon, H; Robb, GR; Sheldon, C; Stokes, S; Morgan, D Identification, optimization, and pharmacology of acylurea GHS-R1a inverse agonists. J Med Chem57:6128-40 (2014) [PubMed] Article McCoull, W; Barton, P; Brown, AJ; Bowker, SS; Cameron, J; Clarke, DS; Davies, RD; Dossetter, AG; Ertan, A; Fenwick, M; Green, C; Holmes, JL; Martin, N; Masters, D; Moore, JE; Newcombe, NJ; Newton, C; Pointon, H; Robb, GR; Sheldon, C; Stokes, S; Morgan, D Identification, optimization, and pharmacology of acylurea GHS-R1a inverse agonists. J Med Chem57:6128-40 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 1/3/4/6/7 |
|---|
| Name: | Histone deacetylase 1/3/4/6/7 |
|---|
| Synonyms: | Histone deacetylase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 1450204 |
|---|
| Components: | This complex has 7 components. |
|---|
| Component 1 |
| Name: | Histone deacetylase |
|---|
| Synonyms: | Hdac2 | Hdac2 protein |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 61427.65 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104809 |
|---|
| Residue: | 546 |
|---|
| Sequence: | ALRSRALPAPARRPAPGCPCCEGRRARRLPQPSCLAAGPRRARAAGGGAAARRRRPEPMA
YSQGGGKKKVCYYYDGDIGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKATA
EEMTKYHSDEYIKFLRSIRPDNMSEYSKQMQRFNVGEDCPVFDGLFEFCQLSTGGSVAGA
VKLNRQQTDMAVNWAGGLHHAKKSEASGFCYVNDIVLAVLELLKYHQRVLYIDIDIHHGD
GVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNFPMRDGIDDESYGQIF
KPIISKVMEMYQPSAVVLQCGADSLSGDRLGCFNLTVKGHAKCVEVVKTFNLPLLMLGGG
GYTIRNVARCWTYETAVALDCEIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTPEYMEK
IKQRLFENLRMLPHAPGVQMQAIPEDAVHEDSGDEDGEDPDKRISIRASDKRIACDEEFS
DSEDEGEGGRRNVADHKKGAKKARIEEDKKEAEDKRTDVKEEDKSKDNSGEKTDTKGAKS
EQLNNP
|
|
|
|---|
| Component 2 |
| Name: | Histone deacetylase 4 |
|---|
| Synonyms: | HD4 | HDAC4_RAT | Hdac4 | Histone deacetylase 4 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 118659.39 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104809 |
|---|
| Residue: | 1077 |
|---|
| Sequence: | MSSQSHPDGLSGRDQPVELLNPARVNHMPSTVDVATALPLQVAPAAVPMDLRLDHQFSLP
LEPALREQQLQQELLALKQKQQIQRQILIAEFQRQHEQLSRQHEAQLHEHIKQQQEMLAM
KHQQELLEHQRKLERHRQEQELEKQHREQKLQQLKNKEKGKESAVASTEVKMKLQEFVLN
KKKALAHRNLNHCMSSDPRYWYGKTQHSSLDQSSPPQSGVSASYNHPVLGMYDAKDDFPL
RKTASEPNLKLRSRLKQKVAERRSSPLLRRKDGPVATALKKRPLDVTDSACSSAPGSGPS
SPNSSSGNVSTENGIAPTVPSTPAETSLAHRLVTREGSVAPLPLYTSPSLPNITLGLPAT
GPAAGAAGQQDAERLALPALQQRISLFPGTHLTPYLSTSPLERDGGAAHNPLLQHMVLLE
QPPTQTPLVTGLGALPLHTQSLVGADRVSPSIHKLRQHRPLGRTQSAPLPQNAQALQHLV
IQQQHQQFLEKHKQQFQQQQLHLSKMISKPSEPPRQPESHPEETEEELREHQALLDEPYL
DRLPGQKEPSLAGVQVKQEPIESEEEEVEATREAEPSQRPATEQELLFRQQALLLEQQRI
HQLRNYQASMEAAGIPVSFGSHRPLSRAQSSPASATFPMSVQEPPTKPRFTTGLVYDTLM
LKHQCTCGNTNSHPEHAGRIQSIWSRLQETGLRGKCECIRGRKATLEELQTVHSEAHTLL
YGTNPLNRQKLDSSLTSVFVRLPCGGVGVDSDTIWNEVHSSGAARLAVGCVVELVFKVAT
GELKNGFAVVRPPGHHAEESTPMGFCYFNSVAIAAKLLQQRLNVSKILIVDWDVHHGNGT
QQAFYNDPNVLYMSLHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLDPPMGDAEY
LAAFRTVVMPIANEFAPDVVLVSSGFDAVEGHPTPLGGYNLSAKCFGYLTKQLMGLAGGR
IVLALEGGHDLTAICDASEACVSALLGNELEPLPEKVLHQRPNANAVHSMEKVMGIHSEY
WRCLQRLSPTVGHSLIEAQKCENEEAETVTAMASLSVGVKPAEKRSEEEPMEEEPPL
|
|
|
|---|
| Component 3 |
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3_RAT | Hdac3 | Histone deacetylase 3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 48802.52 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104809 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
SQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| Component 4 |
| Name: | Histone deacetylase 6 |
|---|
| Synonyms: | Hdac6 | Histone deacetylase 6 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 16154.23 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104809 |
|---|
| Residue: | 158 |
|---|
| Sequence: | ETVGGATTDQCASVAATENSANQTTSGEEASGETESFGTSPSSNASKQTTGASPLHGAAA
QQSPELGLSSTLELSSEAQEVQESEEGLLGEAAGGQDMNSLMLTQGFGDFNTQDVFYAVT
PLSWCPHLMAVCPIPAAGLDVSQPCKTCGSFQENWVCL
|
|
|
|---|
| Component 5 |
| Name: | Histone deacetylase 7 |
|---|
| Synonyms: | HD7 | HD7a | HDAC7_RAT | Hdac7 | Hdac7a | Histone deacetylase 7 | Histone deacetylase 7A |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 26111.41 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104809 |
|---|
| Residue: | 238 |
|---|
| Sequence: | TPGSQPQPMDLRVGQRPTVEPPPEPALLTLQHPQRLHRHLFLAGLQQQQRSAEPMRLSMD
PPLPELQGGQQEQELRQLLNKDKSKRSAVASSVVKQKLAEVILKKQQAALERTVHPSSPS
IPYRTLEPLDTEGAARSVLSSFLPPVPSLPTEPPEHFPLRKTVSEPNLKLRYKPKKSLER
RKNPLLRKESAPPSLRRRPAETLGDSSPSSSSTPASGCSSPNDSEHGPNPALGSEADG
|
|
|
|---|
| Component 6 |
| Name: | Histone deacetylase 1 |
|---|
| Synonyms: | HDAC1_RAT | Hdac1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 55080.30 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_472921 |
|---|
| Residue: | 482 |
|---|
| Sequence: | MAQTQGTKRKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKAN
AEEMTKYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGEDCPVFDGLFEFCQLSTGGSVAS
AVKLNKQQTDIAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHG
DGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAI
FKPVMSKVMEMFQPSAVVLQCGSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNLPMLMLGG
GGYTIRNVARCWTYETAVALDTEIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTNEYLE
KIKQRLFENLRMLPHAPGVQMQAIPEDAIPEESGDEDEEDPDKRISICSSDKRIACEEEF
SDSDEEGEGGRKNSSNFKKAKRVKTEDEKEKDPEEKKEVTEEEKTKEEKPEAKGVKEEVK
MA
|
|
|
|---|
| Component 7 |
| Name: | Histone deacetylase |
|---|
| Synonyms: | Hdac5 | Hdac5 protein |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 34754.92 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104809 |
|---|
| Residue: | 328 |
|---|
| Sequence: | MHSSSAVRMAVGCLVELAFKVAAGELKNGFAIIRPPGHHAEESTAMGFCFFNSVAITAKL
LQQKLSVGKVLIVDWDIHHGNGTQQAFYDDPSVLYISLHRYDNGNFFPGSGAPEEVGGGP
GVGYNVNVAWTGGVDPPIGDVEYLTAFRTVVMPIAHEFSPDVVLVSAGFDAVEGHLSPLG
GYSVTARCFGHLTRQLMTLAGGRVVLALEGGHDLTAICDASEACVSALLSVELQPLDEAV
LQQKPSINAVATLEKVIEIQSKHWSCVQRFATGLGCSLREAQTGEKEEAETVSAMALLSM
GAEQAQAAATQEHSPRPAEEPMEQEPAL
|
|
|
|---|
| BDBM50047416 |
|---|
| n/a |
|---|
| Name | BDBM50047416 |
|---|
| Synonyms: | CHEMBL3319215 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H26ClN5O4S2 |
|---|
| Mol. Mass. | 536.067 |
|---|
| SMILES | CN1CCN(CCCS(=O)(=O)c2ccc3nc(NC(=O)NC(=O)c4ccccc4Cl)sc3c2)CC1 |
|---|
| Structure | 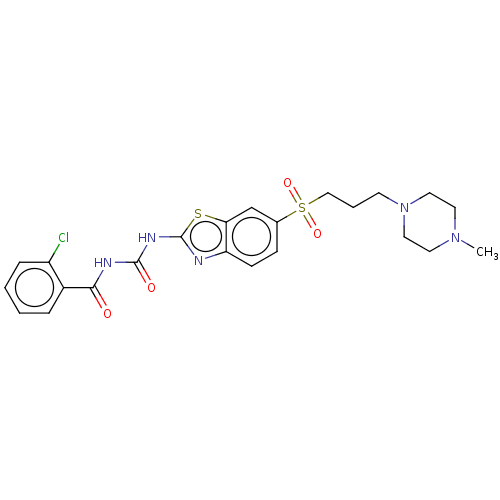
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 McCoull, W; Barton, P; Brown, AJ; Bowker, SS; Cameron, J; Clarke, DS; Davies, RD; Dossetter, AG; Ertan, A; Fenwick, M; Green, C; Holmes, JL; Martin, N; Masters, D; Moore, JE; Newcombe, NJ; Newton, C; Pointon, H; Robb, GR; Sheldon, C; Stokes, S; Morgan, D Identification, optimization, and pharmacology of acylurea GHS-R1a inverse agonists. J Med Chem57:6128-40 (2014) [PubMed] Article
McCoull, W; Barton, P; Brown, AJ; Bowker, SS; Cameron, J; Clarke, DS; Davies, RD; Dossetter, AG; Ertan, A; Fenwick, M; Green, C; Holmes, JL; Martin, N; Masters, D; Moore, JE; Newcombe, NJ; Newton, C; Pointon, H; Robb, GR; Sheldon, C; Stokes, S; Morgan, D Identification, optimization, and pharmacology of acylurea GHS-R1a inverse agonists. J Med Chem57:6128-40 (2014) [PubMed] Article