

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Tyrosine-protein kinase BTK | ||
| Ligand | BDBM50068542 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1466556 (CHEMBL3407013) | ||
| Kd | 6.2±n/a nM | ||
| Citation |  Asami, T; Kawahata, W; Sawa, M TR-FRET binding assay targeting unactivated form of Bruton's tyrosine kinase. Bioorg Med Chem Lett25:2033-6 (2015) [PubMed] Article Asami, T; Kawahata, W; Sawa, M TR-FRET binding assay targeting unactivated form of Bruton's tyrosine kinase. Bioorg Med Chem Lett25:2033-6 (2015) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Tyrosine-protein kinase BTK | |||
| Name: | Tyrosine-protein kinase BTK | ||
| Synonyms: | AGMX1 | ATK | Agammaglobulinaemia tyrosine kinase | Agammaglobulinemia tyrosine kinase | B cell progenitor kinase | B-cell progenitor kinase | BPK | BTK | BTK_HUMAN | Bruton tyrosine kinase | Tyrosine Kinase BTK | Tyrosine-protein kinase (BTK) | Tyrosine-protein kinase BTK (BTK) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 76289.95 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q06187 | ||
| Residue: | 659 | ||
| Sequence: |
| ||
| BDBM50068542 | |||
| n/a | |||
| Name | BDBM50068542 | ||
| Synonyms: | CHEMBL3400826 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C54H49N7O8S | ||
| Mol. Mass. | 956.074 | ||
| SMILES | Cc1c(NC(=O)c2ccc(cc2)C(C)(C)C)cccc1-c1cn(C)c(=O)c(Nc2ccc(cc2)C(=O)NCCCNC(=S)Nc2ccc(c(c2)C(O)=O)-c2c3ccc(O)cc3oc3cc(=O)ccc23)n1 |(-1.18,-23.55,;-1.18,-22.32,;-2.52,-21.55,;-3.85,-22.32,;-5.18,-21.55,;-5.18,-20.32,;-6.52,-22.32,;-7.85,-21.55,;-9.19,-22.32,;-9.18,-23.86,;-7.85,-24.63,;-6.52,-23.86,;-10.52,-24.64,;-11.59,-24.02,;-10.51,-25.87,;-11.58,-25.26,;-2.52,-20.01,;-1.19,-19.24,;.15,-20,;.15,-21.54,;1.49,-22.31,;1.49,-23.85,;2.83,-24.61,;2.83,-25.85,;4.16,-23.84,;5.23,-24.45,;4.16,-22.3,;5.49,-21.52,;5.48,-19.98,;6.81,-19.2,;6.8,-17.66,;5.46,-16.9,;4.13,-17.68,;4.14,-19.22,;5.44,-15.36,;6.51,-14.74,;4.1,-14.6,;4.09,-13.06,;2.75,-12.3,;2.74,-10.76,;1.39,-10,;1.38,-8.46,;2.44,-7.84,;.04,-7.7,;.03,-6.16,;1.35,-5.38,;1.34,-3.84,;0,-3.08,;-1.32,-3.86,;-1.31,-5.4,;-2.66,-3.1,;-2.67,-2.07,;-3.73,-3.73,;,-1.54,;1.31,-.77,;2.67,-1.54,;4,-.77,;4,.77,;5.07,1.39,;2.67,1.54,;1.31,.77,;,1.54,;-1.33,.77,;-2.67,1.54,;-4,.77,;-5.07,1.39,;-4,-.77,;-2.67,-1.54,;-1.33,-.77,;2.82,-21.54,)| | ||
| Structure | 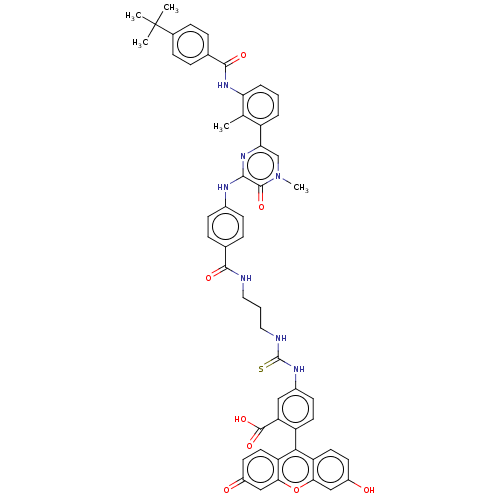
| ||