| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cGMP-dependent 3',5'-cyclic phosphodiesterase |
|---|
| Ligand | BDBM50085603 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1477525 (CHEMBL3430102) |
|---|
| IC50 | 190±n/a nM |
|---|
| Citation |  Rombouts, FJ; Tresadern, G; Buijnsters, P; Langlois, X; Tovar, F; Steinbrecher, TB; Vanhoof, G; Somers, M; Andrés, JI; Trabanco, AA Pyrido[4,3-e][1,2,4]triazolo[4,3-a]pyrazines as Selective, Brain Penetrant Phosphodiesterase 2 (PDE2) Inhibitors. ACS Med Chem Lett6:282-6 (2015) [PubMed] Article Rombouts, FJ; Tresadern, G; Buijnsters, P; Langlois, X; Tovar, F; Steinbrecher, TB; Vanhoof, G; Somers, M; Andrés, JI; Trabanco, AA Pyrido[4,3-e][1,2,4]triazolo[4,3-a]pyrazines as Selective, Brain Penetrant Phosphodiesterase 2 (PDE2) Inhibitors. ACS Med Chem Lett6:282-6 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cGMP-dependent 3',5'-cyclic phosphodiesterase |
|---|
| Name: | cGMP-dependent 3',5'-cyclic phosphodiesterase |
|---|
| Synonyms: | CGS-PDE | Cyclic GMP-stimulated phosphodiesterase | Homo sapiens phosphodiesterase 2A (PDE2A) | NM_002599 | PDE2A | PDE2A_HUMAN | cGSPDE |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 105691.58 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00408 |
|---|
| Residue: | 941 |
|---|
| Sequence: | MGQACGHSILCRSQQYPAARPAEPRGQQVFLKPDEPPPPPQPCADSLQDALLSLGSVIDI
SGLQRAVKEALSAVLPRVETVYTYLLDGESQLVCEDPPHELPQEGKVREAIISQKRLGCN
GLGFSDLPGKPLARLVAPLAPDTQVLVMPLADKEAGAVAAVILVHCGQLSDNEEWSLQAV
EKHTLVALRRVQVLQQRGPREAPRAVQNPPEGTAEDQKGGAAYTDRDRKILQLCGELYDL
DASSLQLKVLQYLQQETRASRCCLLLVSEDNLQLSCKVIGDKVLGEEVSFPLTGCLGQVV
EDKKSIQLKDLTSEDVQQLQSMLGCELQAMLCVPVISRATDQVVALACAFNKLEGDLFTD
EDEHVIQHCFHYTSTVLTSTLAFQKEQKLKCECQALLQVAKNLFTHLDDVSVLLQEIITE
ARNLSNAEICSVFLLDQNELVAKVFDGGVVDDESYEIRIPADQGIAGHVATTGQILNIPD
AYAHPLFYRGVDDSTGFRTRNILCFPIKNENQEVIGVAELVNKINGPWFSKFDEDLATAF
SIYCGISIAHSLLYKKVNEAQYRSHLANEMMMYHMKVSDDEYTKLLHDGIQPVAAIDSNF
ASFTYTPRSLPEDDTSMAILSMLQDMNFINNYKIDCPTLARFCLMVKKGYRDPPYHNWMH
AFSVSHFCYLLYKNLELTNYLEDIEIFALFISCMCHDLDHRGTNNSFQVASKSVLAALYS
SEGSVMERHHFAQAIAILNTHGCNIFDHFSRKDYQRMLDLMRDIILATDLAHHLRIFKDL
QKMAEVGYDRNNKQHHRLLLCLLMTSCDLSDQTKGWKTTRKIAELIYKEFFSQGDLEKAM
GNRPMEMMDREKAYIPELQISFMEHIAMPIYKLLQDLFPKAAELYERVASNREHWTKVSH
KFTIRGLPSNNSLDFLDEEYEVPDLDGTRAPINGCCSLDAE
|
|
|
|---|
| BDBM50085603 |
|---|
| n/a |
|---|
| Name | BDBM50085603 |
|---|
| Synonyms: | CHEMBL3425541 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H10ClN5 |
|---|
| Mol. Mass. | 295.726 |
|---|
| SMILES | Cc1nc2ncccc2n2c(nnc12)-c1ccccc1Cl |
|---|
| Structure | 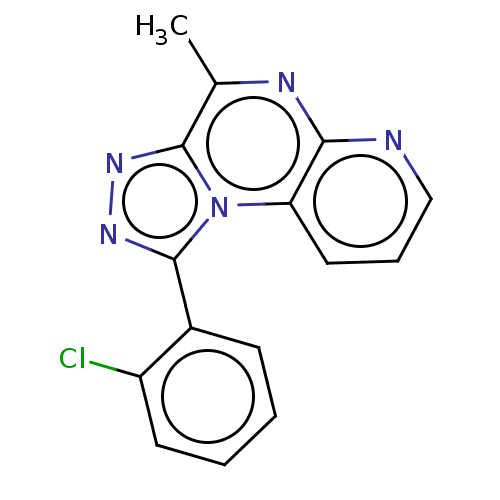
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rombouts, FJ; Tresadern, G; Buijnsters, P; Langlois, X; Tovar, F; Steinbrecher, TB; Vanhoof, G; Somers, M; Andrés, JI; Trabanco, AA Pyrido[4,3-e][1,2,4]triazolo[4,3-a]pyrazines as Selective, Brain Penetrant Phosphodiesterase 2 (PDE2) Inhibitors. ACS Med Chem Lett6:282-6 (2015) [PubMed] Article
Rombouts, FJ; Tresadern, G; Buijnsters, P; Langlois, X; Tovar, F; Steinbrecher, TB; Vanhoof, G; Somers, M; Andrés, JI; Trabanco, AA Pyrido[4,3-e][1,2,4]triazolo[4,3-a]pyrazines as Selective, Brain Penetrant Phosphodiesterase 2 (PDE2) Inhibitors. ACS Med Chem Lett6:282-6 (2015) [PubMed] Article