| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Equilibrative nucleoside transporter 2 |
|---|
| Ligand | BDBM14487 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1494514 (CHEMBL3528991) |
|---|
| Ki | 106000±n/a nM |
|---|
| Citation |  Lepist, EI; Damaraju, VL; Zhang, J; Gati, WP; Yao, SY; Smith, KM; Karpinski, E; Young, JD; Leung, KH; Cass, CE Transport of A1 adenosine receptor agonist tecadenoson by human and mouse nucleoside transporters: evidence for blood-brain barrier transport by murine equilibrative nucleoside transporter 1 mENT1. Drug Metab Dispos41:916-22 (2013) [PubMed] Article Lepist, EI; Damaraju, VL; Zhang, J; Gati, WP; Yao, SY; Smith, KM; Karpinski, E; Young, JD; Leung, KH; Cass, CE Transport of A1 adenosine receptor agonist tecadenoson by human and mouse nucleoside transporters: evidence for blood-brain barrier transport by murine equilibrative nucleoside transporter 1 mENT1. Drug Metab Dispos41:916-22 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Equilibrative nucleoside transporter 2 |
|---|
| Name: | Equilibrative nucleoside transporter 2 |
|---|
| Synonyms: | 36 kDa nucleolar protein HNP36 | DER12 | DER12 | Delayed-early response protein 12 | ENT2 | Equilibrative NBMPR-insensitive nucleoside transporter | Equilibrative nitrobenzylmercaptopurine riboside-insensitive nucleoside transporter | Equilibrative nucleoside transporter 2 | HNP36 | Hydrophobic nucleolar protein, 36 kDa | Nucleoside transporter, ei-type | S29A2_HUMAN | SLC29A2 | Solute carrier family 29 member 2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 50110.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_109695 |
|---|
| Residue: | 456 |
|---|
| Sequence: | MARGDAPRDSYHLVGISFFILGLGTLLPWNFFITAIPYFQARLAGAGNSTARILSTNHTG
PEDAFNFNNWVTLLSQLPLLLFTLLNSFLYQCVPETVRILGSLLAILLLFALTAALVKVD
MSPGPFFSITMASVCFINSFSAVLQGSLFGQLGTMPSTYSTLFLSGQGLAGIFAALAMLL
SMASGVDAETSALGYFITPCVGILMSIVCYLSLPHLKFARYYLANKSSQAQAQELETKAE
LLQSDENGIPSSPQKVALTLDLDLEKEPESEPDEPQKPGKPSVFTVFQKIWLTALCLVLV
FTVTLSVFPAITAMVTSSTSPGKWSQFFNPICCFLLFNIMDWLGRSLTSYFLWPDEDSRL
LPLLVCLRFLFVPLFMLCHVPQRSRLPILFPQDAYFITFMLLFAVSNGYLVSLTMCLAPR
QVLPHEREVAGALMTFFLALGLSCGASLSFLFKALL
|
|
|
|---|
| BDBM14487 |
|---|
| n/a |
|---|
| Name | BDBM14487 |
|---|
| Synonyms: | (2R,3R,4S,5R)-2-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol | Adenine-beta-D-arabinofuranoside | Adenosine | CHEMBL477 | N6-Methylado | [U-14C]adenosine | cid_191 | cid_60961 |
|---|
| Type | Nucleoside or nucleotide |
|---|
| Emp. Form. | C10H13N5O4 |
|---|
| Mol. Mass. | 267.2413 |
|---|
| SMILES | Nc1ncnc2n(cnc12)[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O |
|---|
| Structure | 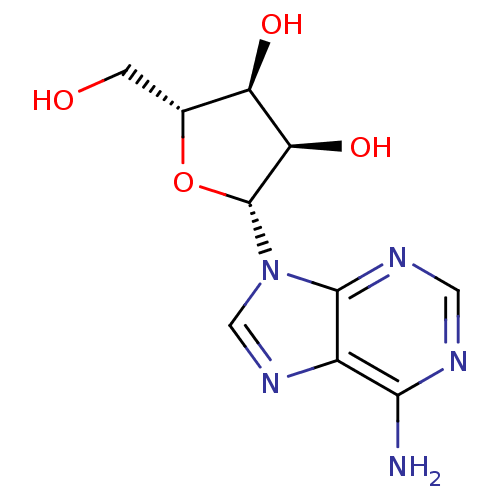
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lepist, EI; Damaraju, VL; Zhang, J; Gati, WP; Yao, SY; Smith, KM; Karpinski, E; Young, JD; Leung, KH; Cass, CE Transport of A1 adenosine receptor agonist tecadenoson by human and mouse nucleoside transporters: evidence for blood-brain barrier transport by murine equilibrative nucleoside transporter 1 mENT1. Drug Metab Dispos41:916-22 (2013) [PubMed] Article
Lepist, EI; Damaraju, VL; Zhang, J; Gati, WP; Yao, SY; Smith, KM; Karpinski, E; Young, JD; Leung, KH; Cass, CE Transport of A1 adenosine receptor agonist tecadenoson by human and mouse nucleoside transporters: evidence for blood-brain barrier transport by murine equilibrative nucleoside transporter 1 mENT1. Drug Metab Dispos41:916-22 (2013) [PubMed] Article