| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prolyl hydroxylase EGLN3 |
|---|
| Ligand | BDBM50149925 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1558377 (CHEMBL3772969) |
|---|
| IC50 | <100000±n/a nM |
|---|
| Citation |  Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Drewes, G; Eagle, R; Garton, N; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Pemberton, M; Rioja, I; Seal, GA; Shipley, T; Singh, O; Suckling, CJ; Taylor, J; Thomas, P; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 2. Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives. J Med Chem59:1370-87 (2016) [PubMed] Article Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Drewes, G; Eagle, R; Garton, N; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Pemberton, M; Rioja, I; Seal, GA; Shipley, T; Singh, O; Suckling, CJ; Taylor, J; Thomas, P; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 2. Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives. J Med Chem59:1370-87 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prolyl hydroxylase EGLN3 |
|---|
| Name: | Prolyl hydroxylase EGLN3 |
|---|
| Synonyms: | EGLN3 | EGLN3_HUMAN | Egl nine homolog 3 (EGLIN3) | Egl nine homolog 3 (EGLN3) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 27265.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9H6Z9 |
|---|
| Residue: | 239 |
|---|
| Sequence: | MPLGHIMRLDLEKIALEYIVPCLHEVGFCYLDNFLGEVVGDCVLERVKQLHCTGALRDGQ
LAGPRAGVSKRHLRGDQITWIGGNEEGCEAISFLLSLIDRLVLYCGSRLGKYYVKERSKA
MVACYPGNGTGYVRHVDNPNGDGRCITCIYYLNKNWDAKLHGGILRIFPEGKSFIADVEP
IFDRLLFFWSDRRNPHEVQPSYATRYAMTVWYFDAEERAEAKKKFRNLTRKTESALTED
|
|
|
|---|
| BDBM50149925 |
|---|
| n/a |
|---|
| Name | BDBM50149925 |
|---|
| Synonyms: | CHEMBL3770740 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H8ClN3O3 |
|---|
| Mol. Mass. | 289.674 |
|---|
| SMILES | Oc1cc(Oc2nc3cnccc3c(=O)[nH]2)ccc1Cl |
|---|
| Structure | 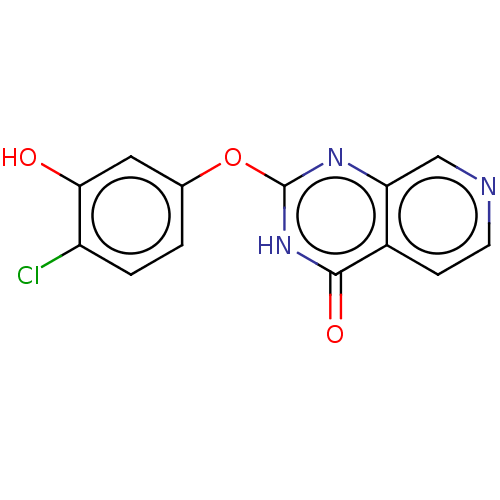
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Drewes, G; Eagle, R; Garton, N; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Pemberton, M; Rioja, I; Seal, GA; Shipley, T; Singh, O; Suckling, CJ; Taylor, J; Thomas, P; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 2. Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives. J Med Chem59:1370-87 (2016) [PubMed] Article
Westaway, SM; Preston, AG; Barker, MD; Brown, F; Brown, JA; Campbell, M; Chung, CW; Drewes, G; Eagle, R; Garton, N; Gordon, L; Haslam, C; Hayhow, TG; Humphreys, PG; Joberty, G; Katso, R; Kruidenier, L; Leveridge, M; Pemberton, M; Rioja, I; Seal, GA; Shipley, T; Singh, O; Suckling, CJ; Taylor, J; Thomas, P; Wilson, DM; Lee, K; Prinjha, RK Cell Penetrant Inhibitors of the KDM4 and KDM5 Families of Histone Lysine Demethylases. 2. Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives. J Med Chem59:1370-87 (2016) [PubMed] Article