| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor ionotropic, NMDA 1/2D |
|---|
| Ligand | BDBM50238367 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1663149 (CHEMBL4012830) |
|---|
| EC50 | 1600±n/a nM |
|---|
| Citation |  Strong, KL; Epplin, MP; Bacsa, J; Butch, CJ; Burger, PB; Menaldino, DS; Traynelis, SF; Liotta, DC The Structure-Activity Relationship of a Tetrahydroisoquinoline Class of N-Methyl-d-Aspartate Receptor Modulators that Potentiates GluN2B-Containing N-Methyl-d-Aspartate Receptors. J Med Chem60:5556-5585 (2017) [PubMed] Article Strong, KL; Epplin, MP; Bacsa, J; Butch, CJ; Burger, PB; Menaldino, DS; Traynelis, SF; Liotta, DC The Structure-Activity Relationship of a Tetrahydroisoquinoline Class of N-Methyl-d-Aspartate Receptor Modulators that Potentiates GluN2B-Containing N-Methyl-d-Aspartate Receptors. J Med Chem60:5556-5585 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor ionotropic, NMDA 1/2D |
|---|
| Name: | Glutamate receptor ionotropic, NMDA 1/2D |
|---|
| Synonyms: | Ionotropic glutamate receptor NMDA 1/2D |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 968983 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Synonyms: | Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Glutamate-NMDA-Channel | Glutamate-NMDA-MK801 | Glutamate-NMDA-Polyamine | Grin1 | NMDA | NMDZ1_RAT | Nmdar1 | phencyclidine |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 105533.40 |
|---|
| Organism: | RAT |
|---|
| Description: | P35439 |
|---|
| Residue: | 938 |
|---|
| Sequence: | MSTMHLLTFALLFSCSFARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQL
NATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLG
LTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYNWNHIILLVSDDHEGRAAQKRL
ETLLEERESKAEKVLQFDPGTKNVTALLMEARELEARVIILSASEDDAATVYRAAAMLNM
TGSGYVWLVGEREISGNALRYAPDGIIGLQLINGKNESAHISDAVGVVAQAVHELLEKEN
ITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRK
LVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTMSDGTC
KEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVA
DGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTI
LVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDA
LTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRP
EERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQA
VRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSH
ENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRH
KDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDT
STGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
|
|
|
|---|
| Component 2 |
| Name: | Glutamate receptor ionotropic, NMDA 2D |
|---|
| Synonyms: | GluN2D | Glutamate [NMDA] receptor subunit epsilon 4 | Grin2d | N-methyl D-aspartate receptor subtype 2D | NMDAR2D | NMDE4_RAT |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 143129.52 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | Q62645 |
|---|
| Residue: | 1323 |
|---|
| Sequence: | MRGAGGPRGPRGPAKMLLLLALACASPFPEEVPGPGAVGGGTGGARPLNVALVFSGPAYA
AEAARLGPAVAAAVRSPGLDVRPVALVLNGSDPRSLVLQLCDLLSGLRVHGVVFEDDSRA
PAVAPILDFLSAQTSLPIVAVHGGAALVLTPKEKGSTFLQLGSSTEQQLQVIFEVLEEYD
WTSFVAVTTRAPGHRAFLSYIEVLTDGSLVGWEHRGALTLDPGAGEAVLGAQLRSVSAQI
RLLFCAREEAEPVFRAAEEAGLTGPGYVWFMVGPQLAGGGGSGVPGEPLLLPGGSPLPAG
LFAVRSAGWRDDLARRVAAGVAVVARGAQALLRDYGFLPELGHDCRTQNRTHRGESLHRY
FMNITWDNRDYSFNEDGFLVNPSLVVISLTRDRTWEVVGSWEQQTLRLKYPLWSRYGRFL
QPVDDTQHLTVATLEERPFVIVEPADPISGTCIRDSVPCRSQLNRTHSPPPDAPRPEKRC
CKGFCIDILKRLAHTIGFSYDLYLVTNGKHGKKIDGVWNGMIGEVFYQRADMAIGSLTIN
EERSEIVDFSVPFVETGISVMVARSNGTVSPSAFLEPYSPAVWVMMFVMCLTVVAVTVFI
FEYLSPVGYNRSLATGKRPGGSTFTIGKSIWLLWALVFNNSVPVENPRGTTSKIMVLVWA
FFAVIFLASYTANLAAFMIQEEYVDTVSGLSDRKFQRPQEQYPPLKFGTVPNGSTEKNIR
SNYPDMHSYMVRYNQPRVEEALTQLKAGKLDAFIYDAAVLNYMARKDEGCKLVTIGSGKV
FATTGYGIALHKGSRWKRPIDLALLQFLGDDEIEMLERLWLSGICHNDKIEVMSSKLDID
NMAGVFYMLLVAMGLSLLVFAWEHLVYWRLRHCLGPTHRMDFLLAFSRGMYSCCSAEAAP
PPAKPPPPPQPLPSPAYPAARPPPGPAPFVPRERAAADRWRRAKGTGPPGGAAIADGFHR
YYGPIEPQGLGLGEARAAPRGAAGRPLSPPTTQPPQKPPPSYFAIVREQEPTEPPAGAFP
GFPSPPAPPAAAAAAVGPPLCRLAFEDESPPAPSRWPRSDPESQPLLGGGAGGPSAGAPT
APPPRRAAPPPCAYLDLEPSPSDSEDSESLGGASLGGLEPWWFADFPYPYAERLGPPPGR
YWSVDKLGGWRAGSWDYLPPRGGPAWHCRHCASLELLPPPRHLSCSHDGLDGGWWAPPPP
PWAAGPPPRRRARCGCPRPHPHRPRASHRAPAAAPHHHRHRRAAGGWDFPPPAPTSRSLE
DLSSCPRAAPTRRLTGPSRHARRCPHAAHWGPPLPTASHRRHRGGDLGTRRGSAHFSSLE
SEV
|
|
|
|---|
| BDBM50238367 |
|---|
| n/a |
|---|
| Name | BDBM50238367 |
|---|
| Synonyms: | CHEMBL4089357 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H30ClNO3S |
|---|
| Mol. Mass. | 496.061 |
|---|
| SMILES | CCOc1ccc(OCC2N(CCc3cc(OC(C)C)ccc23)C(=S)c2cccc(Cl)c2)cc1 |
|---|
| Structure | 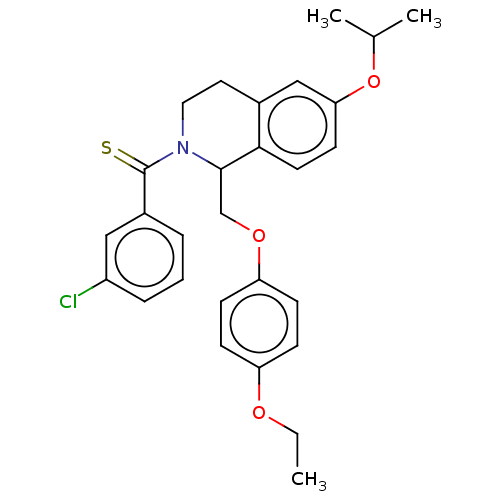
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Strong, KL; Epplin, MP; Bacsa, J; Butch, CJ; Burger, PB; Menaldino, DS; Traynelis, SF; Liotta, DC The Structure-Activity Relationship of a Tetrahydroisoquinoline Class of N-Methyl-d-Aspartate Receptor Modulators that Potentiates GluN2B-Containing N-Methyl-d-Aspartate Receptors. J Med Chem60:5556-5585 (2017) [PubMed] Article
Strong, KL; Epplin, MP; Bacsa, J; Butch, CJ; Burger, PB; Menaldino, DS; Traynelis, SF; Liotta, DC The Structure-Activity Relationship of a Tetrahydroisoquinoline Class of N-Methyl-d-Aspartate Receptor Modulators that Potentiates GluN2B-Containing N-Methyl-d-Aspartate Receptors. J Med Chem60:5556-5585 (2017) [PubMed] Article