| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Ligand | BDBM50022784 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 1000±n/a nM |
|---|
| Comments | PDSP_3689 |
|---|
| Citation |  Bymaster, FP; Katner, JS; Nelson, DL; Hemrick-Luecke, SK; Threlkeld, PG; Heiligenstein, JH; Morin, SM; Gehlert, DR; Perry, KW Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology27:699-711 (2002) [PubMed] Article Bymaster, FP; Katner, JS; Nelson, DL; Hemrick-Luecke, SK; Threlkeld, PG; Heiligenstein, JH; Morin, SM; Gehlert, DR; Perry, KW Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology27:699-711 (2002) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Name: | ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Synonyms: | ATP-regulated potassium channel ROM-K | ATP-sensitive inward rectifier potassium channel 1 | Inward rectifier K(+) channel Kir1.1 | KAB-1 | KCNJ1_RAT | Kcnj1 | Potassium channel (ATP modulatory) | Potassium channel, inwardly rectifying subfamily J member 1 | Renal Outer Medullary Potassium (ROMK) | Renal Outer Medullary Potassium (ROMK1) | Romk1 | The Renal Outer Medullary Potassium (ROMK) channel (Kir1.1) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 44976.27 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P35560 |
|---|
| Residue: | 391 |
|---|
| Sequence: | MGASERSVFRVLIRALTERMFKHLRRWFITHIFGRSRQRARLVSKEGRCNIEFGNVDAQS
RFIFFVDIWTTVLDLKWRYKMTVFITAFLGSWFLFGLLWYVVAYVHKDLPEFYPPDNRTP
CVENINGMTSAFLFSLETQVTIGYGFRFVTEQCATAIFLLIFQSILGVIINSFMCGAILA
KISRPKKRAKTITFSKNAVISKRGGKLCLLIRVANLRKSLLIGSHIYGKLLKTTITPEGE
TIILDQTNINFVVDAGNENLFFISPLTIYHIIDHNSPFFHMAAETLSQQDFELVVFLDGT
VESTSATCQVRTSYVPEEVLWGYRFVPIVSKTKEGKYRVDFHNFGKTVEVETPHCAMCLY
NEKDARARMKRGYDNPNFVLSEVDETDDTQM
|
|
|
|---|
| BDBM50022784 |
|---|
| n/a |
|---|
| Name | BDBM50022784 |
|---|
| Synonyms: | (R)-N-methyl-3-phenyl-3-(o-tolyloxy)propan-1-amine | ATOMOXETINE | CHEMBL641 | Methyl-((R)-3-phenyl-3-o-tolyloxy-propyl)-amine | Methyl-(3-phenyl-3-o-tolyloxy-propyl)-amine | Methyl-(3-phenyl-3-o-tolyloxy-propyl)-amine(tomoxetine) | Strattera | Tomoxetine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H21NO |
|---|
| Mol. Mass. | 255.3547 |
|---|
| SMILES | CNCCC(Oc1ccccc1C)c1ccccc1 |
|---|
| Structure | 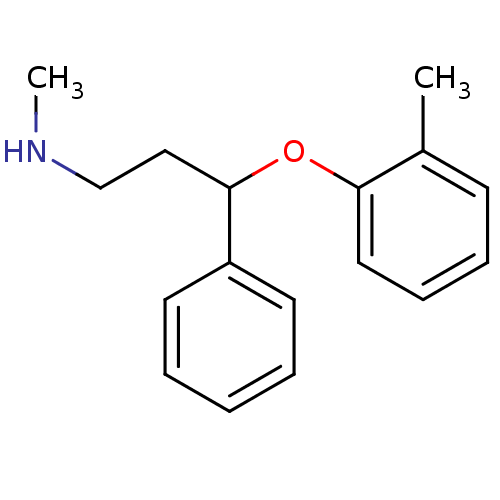
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bymaster, FP; Katner, JS; Nelson, DL; Hemrick-Luecke, SK; Threlkeld, PG; Heiligenstein, JH; Morin, SM; Gehlert, DR; Perry, KW Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology27:699-711 (2002) [PubMed] Article
Bymaster, FP; Katner, JS; Nelson, DL; Hemrick-Luecke, SK; Threlkeld, PG; Heiligenstein, JH; Morin, SM; Gehlert, DR; Perry, KW Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology27:699-711 (2002) [PubMed] Article