| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase FRK |
|---|
| Ligand | BDBM13530 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_6372 |
|---|
| Citation |  Melnick, JS; Janes, J; Kim, S; Chang, JY; Sipes, DG; Gunderson, D; Jarnes, L; Matzen, JT; Garcia, ME; Hood, TL; Beigi, R; Xia, G; Harig, RA; Asatryan, H; Yan, SF; Zhou, Y; Gu, XJ; Saadat, A; Zhou, V; King, FJ; Shaw, CM; Su, AI; Downs, R; Gray, NS; Schultz, PG; Warmuth, M; Caldwell, JS An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A103:3153-8 (2006) [PubMed] Article Melnick, JS; Janes, J; Kim, S; Chang, JY; Sipes, DG; Gunderson, D; Jarnes, L; Matzen, JT; Garcia, ME; Hood, TL; Beigi, R; Xia, G; Harig, RA; Asatryan, H; Yan, SF; Zhou, Y; Gu, XJ; Saadat, A; Zhou, V; King, FJ; Shaw, CM; Su, AI; Downs, R; Gray, NS; Schultz, PG; Warmuth, M; Caldwell, JS An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A103:3153-8 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Tyrosine-protein kinase FRK |
|---|
| Name: | Tyrosine-protein kinase FRK |
|---|
| Synonyms: | FRK | FRK_HUMAN | FYN-related kinase | Nuclear tyrosine protein kinase RAK | PTK5 | RAK | SRC | Tyrosine Kinase FRK |
|---|
| Type: | Tyrosine-protein kinase |
|---|
| Mol. Mass.: | 58254.41 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P42685 |
|---|
| Residue: | 505 |
|---|
| Sequence: | MSNICQRLWEYLEPYLPCLSTEADKSTVIENPGALCSPQSQRHGHYFVALFDYQARTAED
LSFRAGDKLQVLDTLHEGWWFARHLEKRRDGSSQQLQGYIPSNYVAEDRSLQAEPWFFGA
IGRSDAEKQLLYSENKTGSFLIRESESQKGEFSLSVLDGAVVKHYRIKRLDEGGFFLTRR
RIFSTLNEFVSHYTKTSDGLCVKLGKPCLKIQVPAPFDLSYKTVDQWEIDRNSIQLLKRL
GSGQFGEVWEGLWNNTTPVAVKTLKPGSMDPNDFLREAQIMKNLRHPKLIQLYAVCTLED
PIYIITELMRHGSLQEYLQNDTGSKIHLTQQVDMAAQVASGMAYLESRNYIHRDLAARNV
LVGEHNIYKVADFGLARVFKVDNEDIYESRHEIKLPVKWTAPEAIRSNKFSIKSDVWSFG
ILLYEIITYGKMPYSGMTGAQVIQMLAQNYRLPQPSNCPQQFYNIMLECWNAEPKERPTF
ETLRWKLEDYFETDSSYSDANNFIR
|
|
|
|---|
| BDBM13530 |
|---|
| n/a |
|---|
| Name | BDBM13530 |
|---|
| Synonyms: | 4-[(4-methylpiperazin-1-yl)methyl]-N-[4-methyl-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]benzamide | CHEMBL941 | Gleevec | Imatinib | Imatinib, 21 | N-(4-methyl-3-{[4-(pyridin-3-yl)pyrimidin-2-yl]amino}phenyl)-4-[(4-methylpiperazin-1-yl)methyl]benzamide | STI-571 | STI571 | US10906896, Cpd imatinib | US11649218, Example Imatinib | US11725005, Compound imatinib | US9255107, Imatinib | cid_5291 | imatinib-CD3 | med.21724, Compound 6 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H31N7O |
|---|
| Mol. Mass. | 493.6027 |
|---|
| SMILES | CN1CCN(Cc2ccc(cc2)C(=O)Nc2ccc(C)c(Nc3nccc(n3)-c3cccnc3)c2)CC1 |
|---|
| Structure | 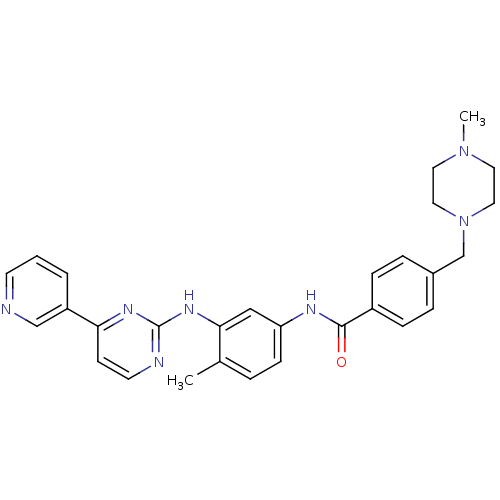
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Melnick, JS; Janes, J; Kim, S; Chang, JY; Sipes, DG; Gunderson, D; Jarnes, L; Matzen, JT; Garcia, ME; Hood, TL; Beigi, R; Xia, G; Harig, RA; Asatryan, H; Yan, SF; Zhou, Y; Gu, XJ; Saadat, A; Zhou, V; King, FJ; Shaw, CM; Su, AI; Downs, R; Gray, NS; Schultz, PG; Warmuth, M; Caldwell, JS An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A103:3153-8 (2006) [PubMed] Article
Melnick, JS; Janes, J; Kim, S; Chang, JY; Sipes, DG; Gunderson, D; Jarnes, L; Matzen, JT; Garcia, ME; Hood, TL; Beigi, R; Xia, G; Harig, RA; Asatryan, H; Yan, SF; Zhou, Y; Gu, XJ; Saadat, A; Zhou, V; King, FJ; Shaw, CM; Su, AI; Downs, R; Gray, NS; Schultz, PG; Warmuth, M; Caldwell, JS An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A103:3153-8 (2006) [PubMed] Article