| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neuronal acetylcholine receptor subunit alpha-4 |
|---|
| Ligand | BDBM86824 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 0.5±n/a nM |
|---|
| Comments | PDSP_8561 |
|---|
| Citation |  Ivy Carroll, F; Yokota, Y; Ma, W; Lee, JR; Brieaddy, LE; Burgess, JP; Navarro, HA; Damaj, MI; Martin, BR Synthesis, nicotinic acetylcholine receptor binding, and pharmacological properties of 3'-(substituted phenyl)deschloroepibatidine analogs. Bioorg Med Chem16:746-54 (2008) [PubMed] Article Ivy Carroll, F; Yokota, Y; Ma, W; Lee, JR; Brieaddy, LE; Burgess, JP; Navarro, HA; Damaj, MI; Martin, BR Synthesis, nicotinic acetylcholine receptor binding, and pharmacological properties of 3'-(substituted phenyl)deschloroepibatidine analogs. Bioorg Med Chem16:746-54 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Neuronal acetylcholine receptor subunit alpha-4 |
|---|
| Name: | Neuronal acetylcholine receptor subunit alpha-4 |
|---|
| Synonyms: | ACHA4_RAT | Acra4 | Cholinergic, Nicotinic Alpha4Beta2 | Cholinergic, Nicotinic Alpha4Beta4 | Chrna4 | Neuronal acetylcholine receptor | Neuronal acetylcholine receptor (alpha4beta2 nAChR) | Neuronal acetylcholine receptor protein alpha-4 subunit | Neuronal acetylcholine receptor subunit alpha 4 beta 2 | Neuronal acetylcholine receptor subunit alpha-4 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 70196.44 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P09483 |
|---|
| Residue: | 630 |
|---|
| Sequence: | MANSGTGAPPPLLLLPLLLLLGTGLLPASSHIETRAHAEERLLKRLFSGYNKWSRPVANI
SDVVLVRFGLSIAQLIDVDEKNQMMTTNVWVKQEWHDYKLRWDPGDYENVTSIRIPSELI
WRPDIVLYNNADGDFAVTHLTKAHLFYDGRVQWTPPAIYKSSCSIDVTFFPFDQQNCTMK
FGSWTYDKAKIDLVSMHSRVDQLDFWESGEWVIVDAVGTYNTRKYECCAEIYPDITYAFI
IRRLPLFYTINLIIPCLLISCLTVLVFYLPSECGEKVTLCISVLLSLTVFLLLITEIIPS
PTSLVIPLIGEYLLFTMIFVTLSIVITVFVLNVHHRSPRTHTMPAWVRRVFLDIVPRLLF
MKRPSVVKDNCRRLIESMHKMANAPRFWPEPVGEPGILSDICNQGLSPAPTFCNPTDTAV
ETQPTCRSPPLEVPDLKTSEVEKASPCPSPGSCPPPKSSSGAPMLIKARSLSVQHVPSSQ
EAAEDGIRCRSRSIQYCVSQDGAASLADSKPTSSPTSLKARPSQLPVSDQASPCKCTCKE
PSPVSPVTVLKAGGTKAPPQHLPLSPALTRAVEGVQYIADHLKAEDTDFSVKEDWKYVAM
VIDRIFLWMFIIVCLLGTVGLFLPPWLAAC
|
|
|
|---|
| BDBM86824 |
|---|
| n/a |
|---|
| Name | BDBM86824 |
|---|
| Synonyms: | CAS_45266048 | NSC_45266048 | rac-3-phenyldeschloroepibatidine dihydrochloride |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H18N2 |
|---|
| Mol. Mass. | 250.3382 |
|---|
| SMILES | C1CC2NC1CC2c1cncc(c1)-c1ccccc1 |TLB:7:6:0.1:3| |
|---|
| Structure | 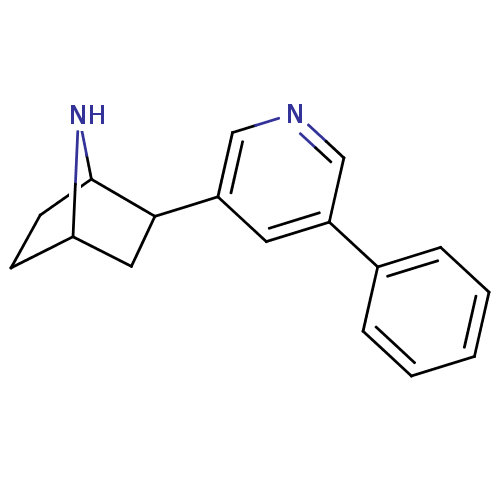
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ivy Carroll, F; Yokota, Y; Ma, W; Lee, JR; Brieaddy, LE; Burgess, JP; Navarro, HA; Damaj, MI; Martin, BR Synthesis, nicotinic acetylcholine receptor binding, and pharmacological properties of 3'-(substituted phenyl)deschloroepibatidine analogs. Bioorg Med Chem16:746-54 (2008) [PubMed] Article
Ivy Carroll, F; Yokota, Y; Ma, W; Lee, JR; Brieaddy, LE; Burgess, JP; Navarro, HA; Damaj, MI; Martin, BR Synthesis, nicotinic acetylcholine receptor binding, and pharmacological properties of 3'-(substituted phenyl)deschloroepibatidine analogs. Bioorg Med Chem16:746-54 (2008) [PubMed] Article