| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2 |
|---|
| Ligand | BDBM50428795 |
|---|
| Substrate/Competitor | n/a |
|---|
| IC50 | 3.9e+3±n/a nM |
|---|
| Citation |  Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2 |
|---|
| Name: | UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2 |
|---|
| Synonyms: | MURA2_STAAM | MurA (S. aureus) | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | murA2 | murZ |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 45068.88 |
|---|
| Organism: | Staphylococcus aureus (Firmicutes) |
|---|
| Description: | S. aureus MurA |
|---|
| Residue: | 419 |
|---|
| Sequence: | MAQEVIKIRGGRTLNGEVNISGAKNSAVAIIPATLLAQGHVKLEGLPQISDVKTLVSLLE
DLNIKASLNGTELEVDTTEIQNAALPNNKVESLRASYYMMGAMLGRFKKCVIGLPGGCPL
GPRPIDQHIKGFKALGAEIDESSTTSMKIEAKELKGAHIFLDMVSVGATINIMLAAVYAT
GQTVIENAAKEPEVVDVANFLTSMGANIKGAGTSTIKINGVKELHGSEYQVIPDRIEAGT
YMCIAAACGENVILNNIVPKHVETLTAKFSELGVNVDVRDERIRINNNAPYQFVDIKTLV
YPGFATDLQQPITPLLFMANGPSFVTDTIYPERFKHVEELKRMGANIEVDEGTATIKPST
LHGAEVYASDLRAGACLIIAGLIAEGVTTIYNVKHIYRGYTDIVEHLKALGADIWTETV
|
|
|
|---|
| BDBM50428795 |
|---|
| n/a |
|---|
| Name | BDBM50428795 |
|---|
| Synonyms: | CHEMBL2335230 | MurA inhibitor (compound 7) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C6H3Br2NO3 |
|---|
| Mol. Mass. | 296.901 |
|---|
| SMILES | [O-][N+](=O)C(\Br)=C\c1ccc(Br)o1 |
|---|
| Structure | 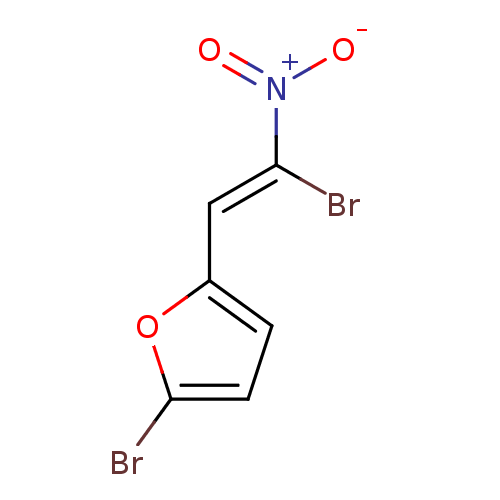
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article
Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article