| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase |
|---|
| Ligand | BDBM119111 |
|---|
| Substrate/Competitor | n/a |
|---|
| IC50 | 7.5e+4±n/a nM |
|---|
| Citation |  Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase |
|---|
| Name: | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase |
|---|
| Synonyms: | MURE_MYCTU | MurE (M. tuberculosis) | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--L-lysine ligase | murE |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55342.10 |
|---|
| Organism: | Mycobacterium tuberculosis H37Rv |
|---|
| Description: | M. tuberculosis MurE |
|---|
| Residue: | 535 |
|---|
| Sequence: | MSSLARGISRRRTEVATQVEAAPTGLRPNAVVGVRLAALADQVGAALAEGPAQRAVTEDR
TVTGVTLRAQDVSPGDLFAALTGSTTHGARHVGDAIARGAVAVLTDPAGVAEIAGRAAVP
VLVHPAPRGVLGGLAATVYGHPSERLTVIGITGTSGKTTTTYLVEAGLRAAGRVAGLIGT
IGIRVGGADLPSALTTPEAPTLQAMLAAMVERGVDTVVMEVSSHALALGRVDGTRFAVGA
FTNLSRDHLDFHPSMADYFEAKASLFDPDSALRARTAVVCIDDDAGRAMAARAADAITVS
AADRPAHWRATDVAPTDAGGQQFTAIDPAGVGHHIGIRLPGRYNVANCLVALAILDTVGV
SPEQAVPGLREIRVPGRLEQIDRGQGFLALVDYAHKPEALRSVLTTLAHPDRRLAVVFGA
GGDRDPGKRAPMGRIAAQLADLVVVTDDNPRDEDPTAIRREILAGAAEVGGDAQVVEIAD
RRDAIRHAVAWARPGDVVLIAGKGHETGQRGGGRVRPFDDRVELAAALEALERRA
|
|
|
|---|
| BDBM119111 |
|---|
| n/a |
|---|
| Name | BDBM119111 |
|---|
| Synonyms: | MurE inhibitor (compound 53) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H28O4 |
|---|
| Mol. Mass. | 332.4339 |
|---|
| SMILES | [#6]-[#6](-[#6])-[#6](=O)-c1c(-[#8])cc(-[#8])cc1-[#8]-[#6]\[#6]=[#6](\[#6])-[#6]-[#6]\[#6]=[#6](\[#6])-[#6] |
|---|
| Structure | 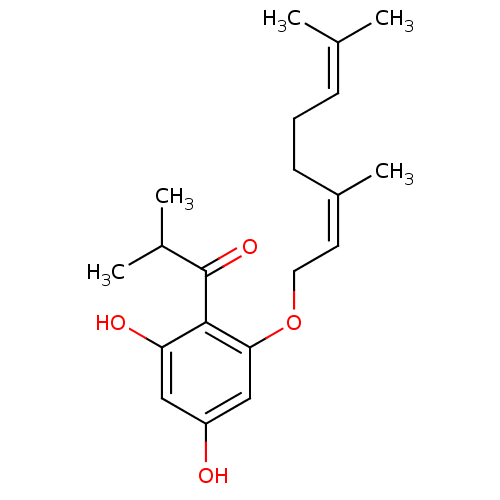
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article
Hrast, M; Sosic, I; Sink, R; Gobec, S Inhibitors of the peptidoglycan biosynthesis enzymes MurA-F. Bioorg Chem55:2-15 (2014) [PubMed] Article