| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cathepsin H |
|---|
| Ligand | BDBM120823 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enzymatic activity assay |
|---|
| pH | 6±n/a |
|---|
| Temperature | 310.15±n/a K |
|---|
| Ki | 1.37±0.132 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Raghav, N; Garg, S N-formylpyrazolines and N-benzoylpyrazolines as novel inhibitors of mammalian cathepsin B and cathepsin H. Bioorg Chem57:43-50 (2014) [PubMed] Article Raghav, N; Garg, S N-formylpyrazolines and N-benzoylpyrazolines as novel inhibitors of mammalian cathepsin B and cathepsin H. Bioorg Chem57:43-50 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cathepsin H |
|---|
| Name: | Cathepsin H |
|---|
| Synonyms: | n/a |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 37394.24 |
|---|
| Organism: | Capra hircus (Goat) |
|---|
| Description: | A0A060INS8 |
|---|
| Residue: | 335 |
|---|
| Sequence: | MWAVLPLLCAGAWLLGAPACGAAELAVNSLEKFHFQSWMVQHQKKYSSEEYHHRLQVFAS
NLREINAHNARNHTFKMGLNQFSDMSFAELKRKYLWSEPQNCSATKSNYLRGTGPYPPSM
DWREKGNFVTPVKNQGSCGSCWTFSTTGALESAVAIATGKLPFLAEQQLVDCAQNFNNHG
CQGGLPSQAFEYIRYNKGIMGEDTYPYRGQDGDCKYQPSKAIAFVKDVANITLNDEEAMV
EAVALYNPVSFAFEVTADFMMYRKGIYSSTSCHKTPDKVNHAVLAVGYGEEKGIPYWIVK
NSWGPHWGMKGYFLIERGKNMCGLAACASFPIPLV
|
|
|
|---|
| BDBM120823 |
|---|
| n/a |
|---|
| Name | BDBM120823 |
|---|
| Synonyms: | N-formyl-5-(-4'-chlorophenyl)-3-phenylpyrazoline (1d) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H13ClN2O |
|---|
| Mol. Mass. | 284.74 |
|---|
| SMILES | Clc1ccc(cc1)C1CC(=NN1C=O)c1ccccc1 |c:10| |
|---|
| Structure | 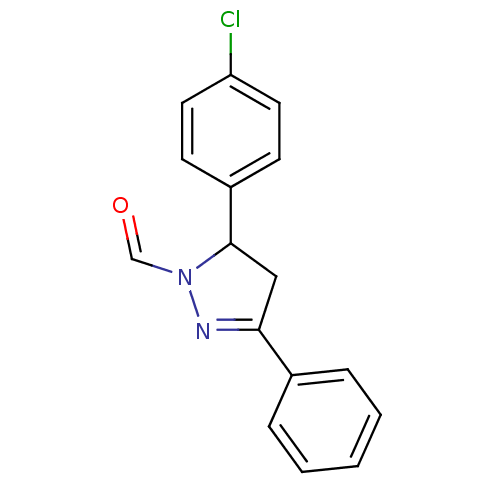
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Raghav, N; Garg, S N-formylpyrazolines and N-benzoylpyrazolines as novel inhibitors of mammalian cathepsin B and cathepsin H. Bioorg Chem57:43-50 (2014) [PubMed] Article
Raghav, N; Garg, S N-formylpyrazolines and N-benzoylpyrazolines as novel inhibitors of mammalian cathepsin B and cathepsin H. Bioorg Chem57:43-50 (2014) [PubMed] Article