| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Nitric oxide synthase, brain |
|---|
| Ligand | BDBM130383 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Spectral Binding Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 298.15±n/a K |
|---|
| Kd | 198± 18 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Li, H; Jamal, J; Delker, S; Plaza, C; Ji, H; Jing, Q; Huang, H; Kang, S; Silverman, RB; Poulos, TL The mobility of a conserved tyrosine residue controls isoform-dependent enzyme-inhibitor interactions in nitric oxide synthases. Biochemistry53:5272-9 (2014) [PubMed] Article Li, H; Jamal, J; Delker, S; Plaza, C; Ji, H; Jing, Q; Huang, H; Kang, S; Silverman, RB; Poulos, TL The mobility of a conserved tyrosine residue controls isoform-dependent enzyme-inhibitor interactions in nitric oxide synthases. Biochemistry53:5272-9 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Nitric oxide synthase, brain |
|---|
| Name: | Nitric oxide synthase, brain |
|---|
| Synonyms: | Bnos | N-NOS | NC-NOS | NOS | NOS type I nNOS | NOS1_RAT | Neuronal nitric oxide synthase | Neuronal nitric oxide synthase (nNOS) | Nitric Oxide Synthase, brain | Nitric oxide synthase (nNOS) | Nitric oxide synthase, brain (nNOS) | Nitric-oxide synthase, brain | Nitric-oxide synthase, brain (nNOS) | Nitrogen oxide synthase - neuronal | Nos1 | Peptidyl-cysteine S-nitrosylase NOS1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 160570.98 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | Recombinant nNOS overexpressed in E. coli was used in enzyme assays. |
|---|
| Residue: | 1429 |
|---|
| Sequence: | MEENTFGVQQIQPNVISVRLFKRKVGGLGFLVKERVSKPPVIISDLIRGGAAEQSGLIQA
GDIILAVNDRPLVDLSYDSALEVLRGIASETHVVLILRGPEGFTTHLETTFTGDGTPKTI
RVTQPLGPPTKAVDLSHQPSASKDQSLAVDRVTGLGNGPQHAQGHGQGAGSVSQANGVAI
DPTMKSTKANLQDIGEHDELLKEIEPVLSILNSGSKATNRGGPAKAEMKDTGIQVDRDLD
GKSHKAPPLGGDNDRVFNDLWGKDNVPVILNNPYSEKEQSPTSGKQSPTKNGSPSRCPRF
LKVKNWETDVVLTDTLHLKSTLETGCTEHICMGSIMLPSQHTRKPEDVRTKDQLFPLAKE
FLDQYYSSIKRFGSKAHMDRLEEVNKEIESTSTYQLKDTELIYGAKHAWRNASRCVGRIQ
WSKLQVFDARDCTTAHGMFNYICNHVKYATNKGNLRSAITIFPQRTDGKHDFRVWNSQLI
RYAGYKQPDGSTLGDPANVQFTEICIQQGWKAPRGRFDVLPLLLQANGNDPELFQIPPEL
VLEVPIRHPKFDWFKDLGLKWYGLPAVSNMLLEIGGLEFSACPFSGWYMGTEIGVRDYCD
NSRYNILEEVAKKMDLDMRKTSSLWKDQALVEINIAVLYSFQSDKVTIVDHHSATESFIK
HMENEYRCRGGCPADWVWIVPPMSGSITPVFHQEMLNYRLTPSFEYQPDPWNTHVWKGTN
GTPTKRRAIGFKKLAEAVKFSAKLMGQAMAKRVKATILYATETGKSQAYAKTLCEIFKHA
FDAKAMSMEEYDIVHLEHEALVLVVTSTFGNGDPPENGEKFGCALMEMRHPNSVQEERKS
YKVRFNSVSSYSDSRKSSGDGPDLRDNFESTGPLANVRFSVFGLGSRAYPHFCAFGHAVD
TLLEELGGERILKMREGDELCGQEEAFRTWAKKVFKAACDVFCVGDDVNIEKPNNSLISN
DRSWKRNKFRLTYVAEAPDLTQGLSNVHKKRVSAARLLSRQNLQSPKFSRSTIFVRLHTN
GNQELQYQPGDHLGVFPGNHEDLVNALIERLEDAPPANHVVKVEMLEERNTALGVISNWK
DESRLPPCTIFQAFKYYLDITTPPTPLQLQQFASLATNEKEKQRLLVLSKGLQEYEEWKW
GKNPTMVEVLEEFPSIQMPATLLLTQLSLLQPRYYSISSSPDMYPDEVHLTVAIVSYHTR
DGEGPVHHGVCSSWLNRIQADDVVPCFVRGAPSFHLPRNPQVPCILVGPGTGIAPFRSFW
QQRQFDIQHKGMNPCPMVLVFGCRQSKIDHIYREETLQAKNKGVFRELYTAYSREPDRPK
KYVQDVLQEQLAESVYRALKEQGGHIYVCGDVTMAADVLKAIQRIMTQQGKLSEEDAGVF
ISRLRDDNRYHEDIFGVTLRTYEVTNRLRSESIAFIEESKKDADEVFSS
|
|
|
|---|
| BDBM130383 |
|---|
| n/a |
|---|
| Name | BDBM130383 |
|---|
| Synonyms: | 4methyl6{[(3R,4R)4{[5(4methylpyridin2 yl)pentyl]oxy}pyrrolidin3yl]methyl}pyridin2amine ((3R,4R)-3) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H32N4O |
|---|
| Mol. Mass. | 368.5157 |
|---|
| SMILES | Cc1ccnc(CCCCCO[C@H]2CNC[C@H]2Cc2cc(C)cc(N)n2)c1 |r| |
|---|
| Structure | 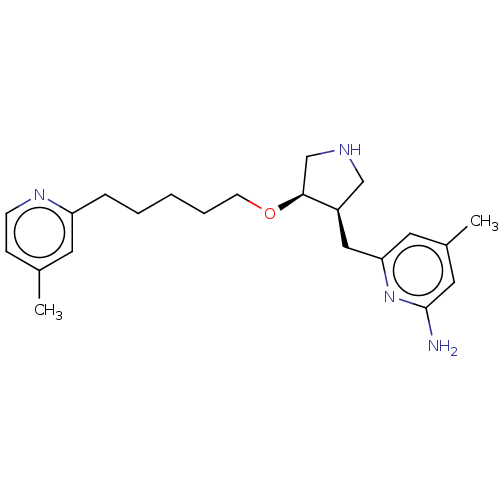
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, H; Jamal, J; Delker, S; Plaza, C; Ji, H; Jing, Q; Huang, H; Kang, S; Silverman, RB; Poulos, TL The mobility of a conserved tyrosine residue controls isoform-dependent enzyme-inhibitor interactions in nitric oxide synthases. Biochemistry53:5272-9 (2014) [PubMed] Article
Li, H; Jamal, J; Delker, S; Plaza, C; Ji, H; Jing, Q; Huang, H; Kang, S; Silverman, RB; Poulos, TL The mobility of a conserved tyrosine residue controls isoform-dependent enzyme-inhibitor interactions in nitric oxide synthases. Biochemistry53:5272-9 (2014) [PubMed] Article