| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 5D [1-760] |
|---|
| Ligand | BDBM191604 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Formaldehyde Dehydrogenase-Coupled Demethylase (FDH) Assay |
|---|
| Temperature | 298.15±n/a K |
|---|
| IC50 | >2.00e+5±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Horton, JR; Liu, X; Gale, M; Wu, L; Shanks, JR; Zhang, X; Webber, PJ; Bell, JS; Kales, SC; Mott, BT; Rai, G; Jansen, DJ; Henderson, MJ; Urban, DJ; Hall, MD; Simeonov, A; Maloney, DJ; Johns, MA; Fu, H; Jadhav, A; Vertino, PM; Yan, Q; Cheng, X Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem Biol23:769-81 (2016) [PubMed] Article Horton, JR; Liu, X; Gale, M; Wu, L; Shanks, JR; Zhang, X; Webber, PJ; Bell, JS; Kales, SC; Mott, BT; Rai, G; Jansen, DJ; Henderson, MJ; Urban, DJ; Hall, MD; Simeonov, A; Maloney, DJ; Johns, MA; Fu, H; Jadhav, A; Vertino, PM; Yan, Q; Cheng, X Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem Biol23:769-81 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 5D [1-760] |
|---|
| Name: | Lysine-specific demethylase 5D [1-760] |
|---|
| Synonyms: | HY | HYA | JARID1D | KDM5D | KDM5D_HUMAN | KIAA0234 | Lysine-specific demethylase 5D (KDM5D(aa 1-760)-AP) | SMCY |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 87351.60 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Truncation 1-760 aa. Contains deletion of ARID and PhD1 domains. |
|---|
| Residue: | 760 |
|---|
| Sequence: | MEPGCDEFLPPPECPVFEPSWAEFQDPLGYIAKIRPIAEKSGICKIRPPADWQPPFAVEV
DNFRFTPRVQRLNELEAQTRVKLNYLDQIAKFWEIQGSSLKIPNVERKILDLYSLSKIVI
EEGGYEAICKDRRWARVAQRLHYPPGKNIGSLLRSHYERIIYPYEMFQSGANHVQCNTHP
FDNEVKDKEYKPHSIPLRQSVQPSKFSSYSRRAKRLQPDPEPTEEDIEKHPELKKLQIYG
PGPKMMGLGLMAKDKDKTVHKKVTCPPTVTVKDEQSGGGNVSSTLLKQHLSLEPCTKTTM
QLRKNHSSAQFIDSYICQVCSRGDEDDKLLFCDGCDDNYHIFCLLPPLPEIPRGIWRCPK
CILAECKQPPEAFGFEQATQEYSLQSFGEMADSFKSDYFNMPVHMVPTELVEKEFWRLVS
SIEEDVTVEYGADIHSKEFGSGFPVSNSKQNLSPEEKEYATSGWNLNVMPVLDQSVLCHI
NADISGMKVPWLYVGMVFSAFCWHIEDHWSYSINYLHWGEPKTWYGVPSLAAEHLEEVMK
MLTPELFDSQPDLLHQLVTLMNPNTLMSHGVPVVRTNQCAGEFVITFPRAYHSGFNQGYN
FAEAVNFCTADWLPAGRQCIEHYRRLRRYCVFSHEELICKMAAFPETLDLNLAVAVHKEM
FIMVQEERRLRKALLEKGVTEAEREAFELLPDDERQCIKCKTTCFLSALACYDCPDGLVC
LSHINDLCKCSSSRQYLRYRYTLDELPTMLHKLKIRAESF
|
|
|
|---|
| BDBM191604 |
|---|
| n/a |
|---|
| Name | BDBM191604 |
|---|
| Synonyms: | 8-(1-methyl-1Himidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol (N10) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H14F3N5O2 |
|---|
| Mol. Mass. | 353.2992 |
|---|
| SMILES | Cn1cnc(c1)-c1nccc2c(O)nc(OCCCC(F)(F)F)nc12 |
|---|
| Structure | 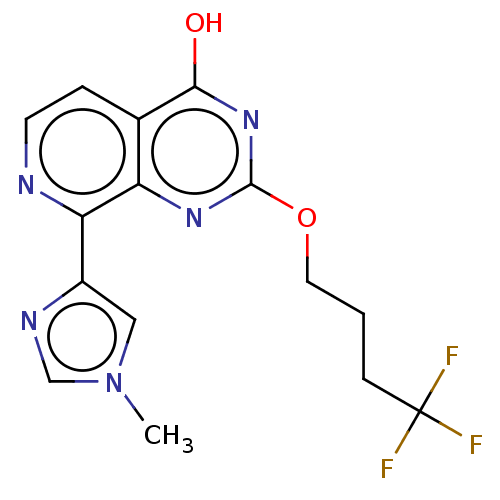
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Horton, JR; Liu, X; Gale, M; Wu, L; Shanks, JR; Zhang, X; Webber, PJ; Bell, JS; Kales, SC; Mott, BT; Rai, G; Jansen, DJ; Henderson, MJ; Urban, DJ; Hall, MD; Simeonov, A; Maloney, DJ; Johns, MA; Fu, H; Jadhav, A; Vertino, PM; Yan, Q; Cheng, X Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem Biol23:769-81 (2016) [PubMed] Article
Horton, JR; Liu, X; Gale, M; Wu, L; Shanks, JR; Zhang, X; Webber, PJ; Bell, JS; Kales, SC; Mott, BT; Rai, G; Jansen, DJ; Henderson, MJ; Urban, DJ; Hall, MD; Simeonov, A; Maloney, DJ; Johns, MA; Fu, H; Jadhav, A; Vertino, PM; Yan, Q; Cheng, X Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem Biol23:769-81 (2016) [PubMed] Article