| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 8 |
|---|
| Ligand | BDBM25150 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | In vitro HDACs Inhibition Fluorescence Assay |
|---|
| IC50 | 353±49 nM |
|---|
| Citation |  Li, J; Li, X; Wang, X; Hou, J; Zang, J; Gao, S; Xu, W; Zhang, Y PXD101 analogs with L-phenylglycine-containing branched cap as histone deacetylase inhibitors. Chem Biol Drug Des88:574-84 (2016) [PubMed] Article Li, J; Li, X; Wang, X; Hou, J; Zang, J; Gao, S; Xu, W; Zhang, Y PXD101 analogs with L-phenylglycine-containing branched cap as histone deacetylase inhibitors. Chem Biol Drug Des88:574-84 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 8 |
|---|
| Name: | Histone deacetylase 8 |
|---|
| Synonyms: | HD8 | HDAC8 | HDAC8_HUMAN | HDACL1 | Histone deacetylase 8 (HDAC-8) | Human HDAC8 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 41749.60 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9BY41 |
|---|
| Residue: | 377 |
|---|
| Sequence: | MEEPEEPADSGQSLVPVYIYSPEYVSMCDSLAKIPKRASMVHSLIEAYALHKQMRIVKPK
VASMEEMATFHTDAYLQHLQKVSQEGDDDHPDSIEYGLGYDCPATEGIFDYAAAIGGATI
TAAQCLIDGMCKVAINWSGGWHHAKKDEASGFCYLNDAVLGILRLRRKFERILYVDLDLH
HGDGVEDAFSFTSKVMTVSLHKFSPGFFPGTGDVSDVGLGKGRYYSVNVPIQDGIQDEKY
YQICESVLKEVYQAFNPKAVVLQLGADTIAGDPMCSFNMTPVGIGKCLKYILQWQLATLI
LGGGGYNLANTARCWTYLTGVILGKTLSSEIPDHEFFTAYGPDYVLEITPSCRPDRNEPH
RIQQILNYIKGNLKHVV
|
|
|
|---|
| BDBM25150 |
|---|
| n/a |
|---|
| Name | BDBM25150 |
|---|
| Synonyms: | (2E)-N-hydroxy-3-[3-(phenylsulfamoyl)phenyl]prop-2-enamide | Belinosta | Belinostat | PXD-101 | PXD101 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H14N2O4S |
|---|
| Mol. Mass. | 318.348 |
|---|
| SMILES | ONC(=O)\C=C\c1cccc(c1)S(=O)(=O)Nc1ccccc1 |
|---|
| Structure | 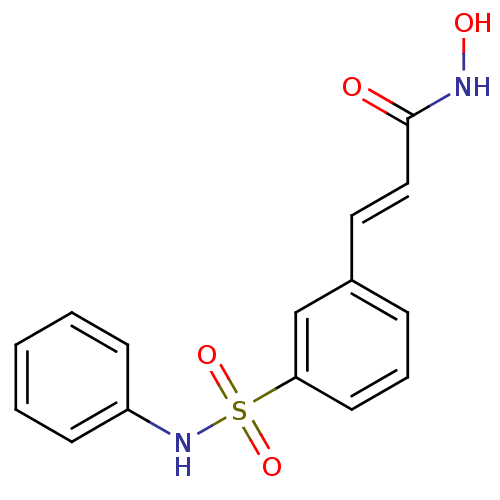
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, J; Li, X; Wang, X; Hou, J; Zang, J; Gao, S; Xu, W; Zhang, Y PXD101 analogs with L-phenylglycine-containing branched cap as histone deacetylase inhibitors. Chem Biol Drug Des88:574-84 (2016) [PubMed] Article
Li, J; Li, X; Wang, X; Hou, J; Zang, J; Gao, S; Xu, W; Zhang, Y PXD101 analogs with L-phenylglycine-containing branched cap as histone deacetylase inhibitors. Chem Biol Drug Des88:574-84 (2016) [PubMed] Article