| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Seed linoleate 13S-lipoxygenase-1 |
|---|
| Ligand | BDBM222271 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | In Vitro Soybean Lipoxygenase (SLO) Inhibition Assay |
|---|
| pH | 8±n/a |
|---|
| IC50 | 2.26e+4± 3.0 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Vinayagam, J; Gajbhiye, RL; Mandal, L; Arumugam, M; Achari, A; Jaisankar, P Substituted furans as potent lipoxygenase inhibitors: Synthesis, in vitro and molecular docking studies. Bioorg Chem71:97-101 (2017) [PubMed] Article Vinayagam, J; Gajbhiye, RL; Mandal, L; Arumugam, M; Achari, A; Jaisankar, P Substituted furans as potent lipoxygenase inhibitors: Synthesis, in vitro and molecular docking studies. Bioorg Chem71:97-101 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Seed linoleate 13S-lipoxygenase-1 |
|---|
| Name: | Seed linoleate 13S-lipoxygenase-1 |
|---|
| Synonyms: | 15-LOX | 15-Lipo-oxygenase (15-LO) | 15-lipo-oxygenase (SLO) | Arachidonic Acid 15- Lipoxygenase | L-1 | LOX1 | LOX1.1 | LOX1_SOYBN | Lipoxygenase (LOX) | Lipoxygenase (SLO) | Lipoxygenase-1 | Seed lipoxygenase-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 94365.66 |
|---|
| Organism: | Glycine max (soybean) |
|---|
| Description: | n/a |
|---|
| Residue: | 839 |
|---|
| Sequence: | MFSAGHKIKGTVVLMPKNELEVNPDGSAVDNLNAFLGRSVSLQLISATKADAHGKGKVGK
DTFLEGINTSLPTLGAGESAFNIHFEWDGSMGIPGAFYIKNYMQVEFFLKSLTLEAISNQ
GTIRFVCNSWVYNTKLYKSVRIFFANHTYVPSETPAPLVSYREEELKSLRGNGTGERKEY
DRIYDYDVYNDLGNPDKSEKLARPVLGGSSTFPYPRRGRTGRGPTVTDPNTEKQGEVFYV
PRDENLGHLKSKDALEIGTKSLSQIVQPAFESAFDLKSTPIEFHSFQDVHDLYEGGIKLP
RDVISTIIPLPVIKELYRTDGQHILKFPQPHVVQVSQSAWMTDEEFAREMIAGVNPCVIR
GLEEFPPKSNLDPAIYGDQSSKITADSLDLDGYTMDEALGSRRLFMLDYHDIFMPYVRQI
NQLNSAKTYATRTILFLREDGTLKPVAIELSLPHSAGDLSAAVSQVVLPAKEGVESTIWL
LAKAYVIVNDSCYHQLMSHWLNTHAAMEPFVIATHRHLSVLHPIYKLLTPHYRNNMNINA
LARQSLINANGIIETTFLPSKYSVEMSSAVYKNWVFTDQALPADLIKRGVAIKDPSTPHG
VRLLIEDYPYAADGLEIWAAIKTWVQEYVPLYYARDDDVKNDSELQHWWKEAVEKGHGDL
KDKPWWPKLQTLEDLVEVCLIIIWIASALHAAVNFGQYPYGGLIMNRPTASRRLLPEKGT
PEYEEMINNHEKAYLRTITSKLPTLISLSVIEILSTHASDEVYLGQRDNPHWTSDSKALQ
AFQKFGNKLKEIEEKLVRRNNDPSLQGNRLGPVQLPYTLLYPSSEEGLTFRGIPNSISI
|
|
|
|---|
| BDBM222271 |
|---|
| n/a |
|---|
| Name | BDBM222271 |
|---|
| Synonyms: | 2-Methyl-4-(2-oxo-2-p-tolyl-ethyl)-5-p-tolyl-furan-3-carboxylic acid ethyl ester (3d) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H24O4 |
|---|
| Mol. Mass. | 376.445 |
|---|
| SMILES | CCOC(=O)c1c(C)oc(c1CC(=O)c1ccc(C)cc1)-c1ccc(C)cc1 |
|---|
| Structure | 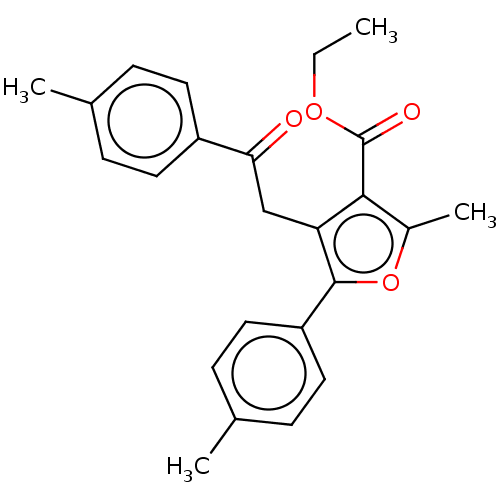
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vinayagam, J; Gajbhiye, RL; Mandal, L; Arumugam, M; Achari, A; Jaisankar, P Substituted furans as potent lipoxygenase inhibitors: Synthesis, in vitro and molecular docking studies. Bioorg Chem71:97-101 (2017) [PubMed] Article
Vinayagam, J; Gajbhiye, RL; Mandal, L; Arumugam, M; Achari, A; Jaisankar, P Substituted furans as potent lipoxygenase inhibitors: Synthesis, in vitro and molecular docking studies. Bioorg Chem71:97-101 (2017) [PubMed] Article