

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipoxygenase (Glycine max (Soybean)) | BDBM222272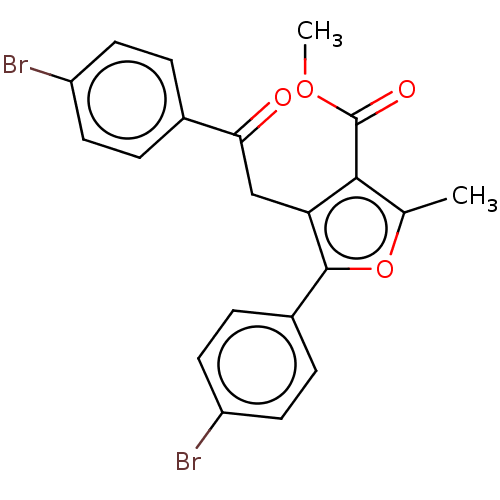 (5-(4-Bromo-phenyl)-4[2-(4-bromo-phenyl)-2-oxo-ethy...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 70.9 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222273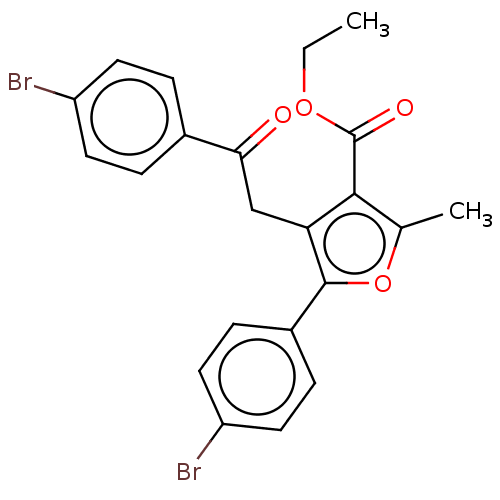 (5-(4-Bromo-phenyl)-4[2-(4-bromo-phenyl)-2-oxo-ethy...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 141 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222270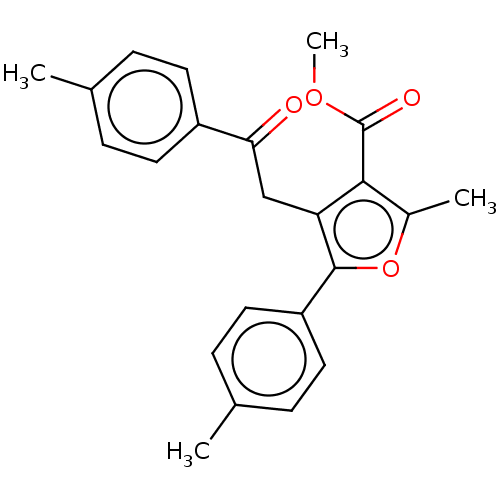 (Methyl 2-methyl-5-(4-methylphenyl)-4-[2-(4-methylp...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 179 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM32020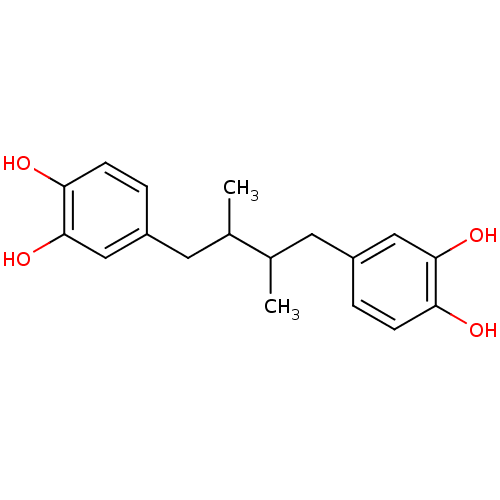 (4-[4-(3,4-dihydroxyphenyl)-2,3-dimethyl-butyl]pyro...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 232 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222271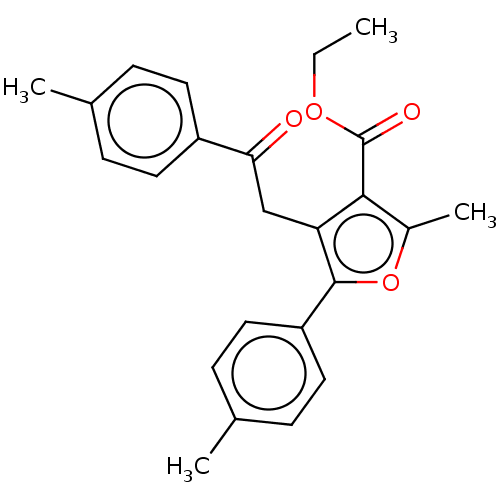 (2-Methyl-4-(2-oxo-2-p-tolyl-ethyl)-5-p-tolyl-furan...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 246 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222274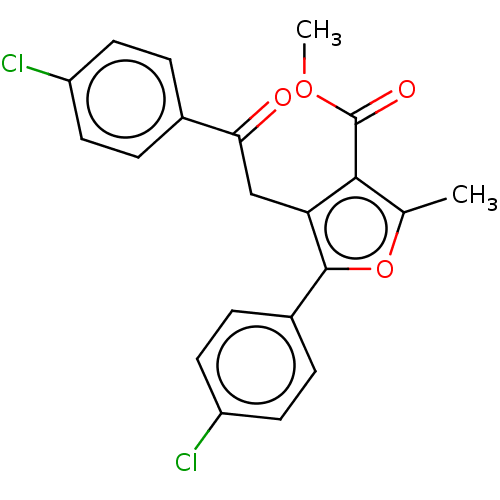 (5-(4-Chloro-phenyl)-4[2-(4-chloro-phenyl)-2-oxo-et...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 313 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222275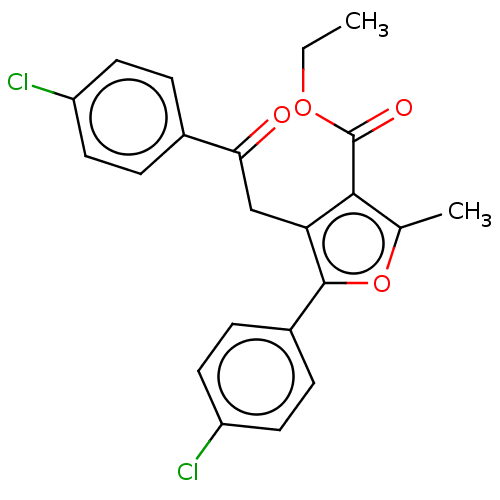 (5-(4-Chloro-phenyl)-4[2-(4-chloro-phenyl)-2-oxo-et...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 345 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222277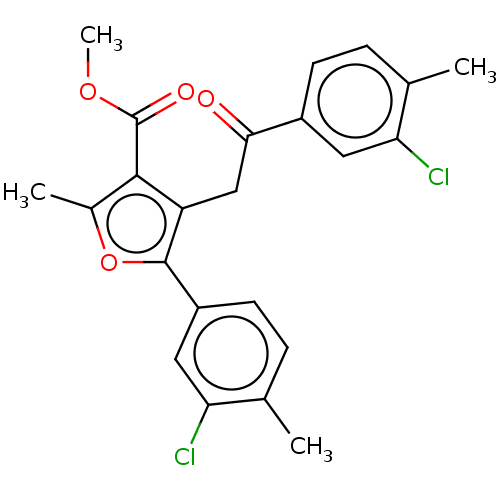 (5-(3-Chloro-4-methyl-phenyl)-4-[2-(3-chloro-4-meth...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 357 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222276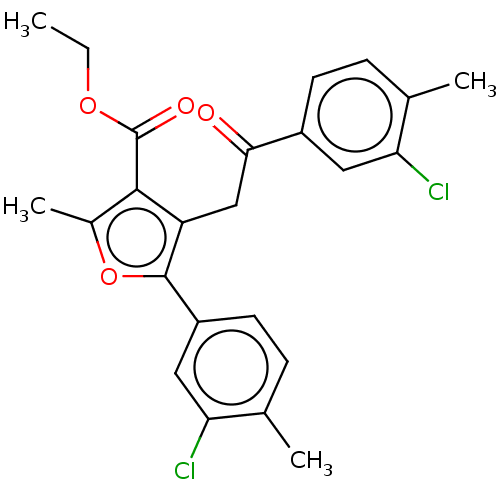 (5-(3-Chloro-4-methyl-phenyl)-4-[2-(3-chloro-4-meth...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 539 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222268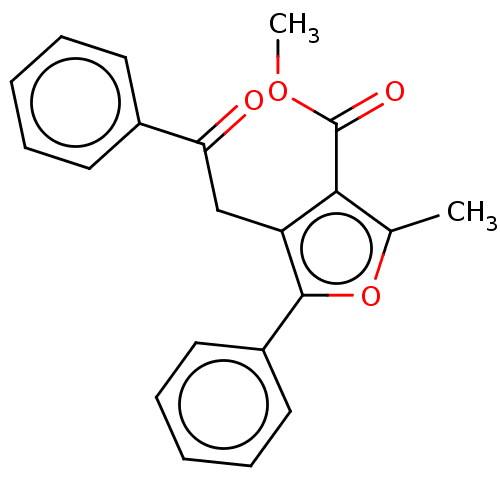 (2-Methyl-4-(2-oxo-2-phenyl-ethyl)-5-phenyl-furan-3...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 662 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lipoxygenase (Glycine max (Soybean)) | BDBM222269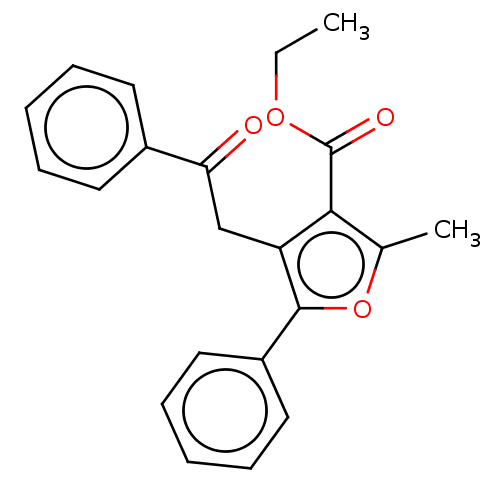 (2-Methyl-4-(2-oxo-2-phenyl-ethyl)-5-phenyl-furan-3...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 868 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description The crystal structure of soybean lipoxygenase-1 (PDB code: 3PZW), was used in our docking experiments. The 3D structure of 3PZW was reported by Chrus... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222272 (5-(4-Bromo-phenyl)-4[2-(4-bromo-phenyl)-2-oxo-ethy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.28E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222273 (5-(4-Bromo-phenyl)-4[2-(4-bromo-phenyl)-2-oxo-ethy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2.12E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222270 (Methyl 2-methyl-5-(4-methylphenyl)-4-[2-(4-methylp...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2.21E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222271 (2-Methyl-4-(2-oxo-2-p-tolyl-ethyl)-5-p-tolyl-furan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2.26E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222275 (5-(4-Chloro-phenyl)-4[2-(4-chloro-phenyl)-2-oxo-et...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 3.20E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222274 (5-(4-Chloro-phenyl)-4[2-(4-chloro-phenyl)-2-oxo-et...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 3.25E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM32020 (4-[4-(3,4-dihydroxyphenyl)-2,3-dimethyl-butyl]pyro...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 3.27E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222277 (5-(3-Chloro-4-methyl-phenyl)-4-[2-(3-chloro-4-meth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 4.75E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222276 (5-(3-Chloro-4-methyl-phenyl)-4-[2-(3-chloro-4-meth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 5.65E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222268 (2-Methyl-4-(2-oxo-2-phenyl-ethyl)-5-phenyl-furan-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 7.85E+4 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222412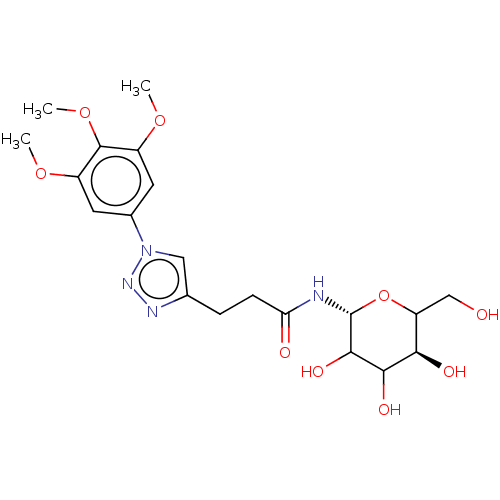 (3-[1-(3,4, 5-trimethoxyphenyl)-1H-[1,2,3]triazol-4...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.21E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222411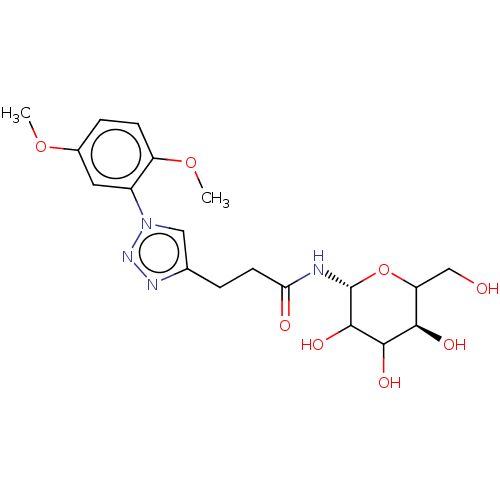 (3-[1-(2,5-Dimethoxyphenyl)-1H-[1,2,3]triazol-4-yl]...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.28E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM23406 ((3R,4R,5S,6R)-5-{[(2R,3R,4R,5S,6R)-5-{[(2R,3R,4S,5...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 1.31E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222407 (3-[1-(4-Hydroxyphenyl)-1H-[1,2,3]triazol-4-yl]-N-(...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.48E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222406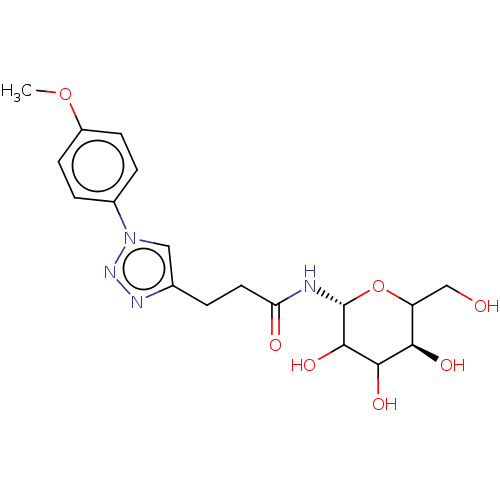 (3-[1-(4-Methoxyphenyl)-1H-[1,2,3]triazol-4-yl]-N-(...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.56E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222408 (3-[1-(4-Methylphenyl)-1H-[1,2,3]triazol-4-yl]-N-(2...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 1.89E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222405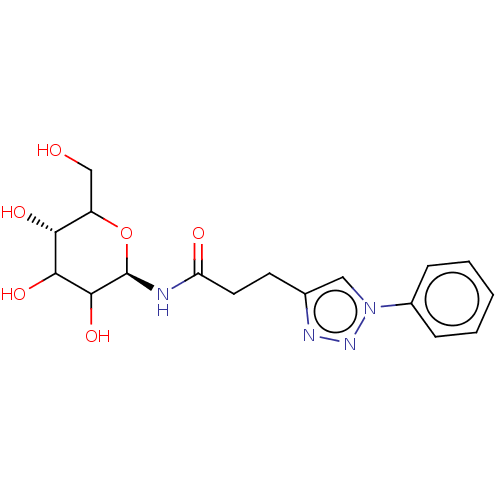 (3-(1-Phenyl-1H-[1,2,3]triazol-4-yl)-N-(2,3,4,6-tet...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2.66E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Seed linoleate 13S-lipoxygenase-1 (Glycine max (soybean)) | BDBM222269 (2-Methyl-4-(2-oxo-2-phenyl-ethyl)-5-phenyl-furan-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2.87E+5 | n/a | n/a | n/a | n/a | 8.0 | n/a |
CSIR - Indian Institute of Chemical Biology | Assay Description SLO inhibition was studied by spectrophotometric assay by measuring the change in the absorbance at 234 nm due to formation of hydroperoxylinoleic ac... | Bioorg Chem 71: 97-101 (2017) Article DOI: 10.1016/j.bioorg.2017.01.016 BindingDB Entry DOI: 10.7270/Q2F76BCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222409 (3-(1-Benzyl-1H-[1,2,3]triazol-4-yl)-N-(2,3,4,6-tet...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 3.42E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL32 (Saccharomyces cerevisiae) | BDBM222410 (3-(1-Naphthyl-1H-[1,2,3]triazol-4-yl)-N-(2,3,4,6-t...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 4.57E+5 | n/a | n/a | n/a | n/a | 6.8 | 1 |
CSIR-Indian Institute of Chemical Biology | Assay Description Briefly, the sample solution (25 μL, 1.0 mM solution in DMSO) was mixed with enzyme solution (25 μL, 0.5 U/mL prepared in 0.1 M phosphate buffe... | Bioorg Chem 72: 11-20 (2017) Article DOI: 10.1016/j.bioorg.2017.03.006 BindingDB Entry DOI: 10.7270/Q2D7999X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA topoisomerase I (Leishmania donovani) | BDBM50239186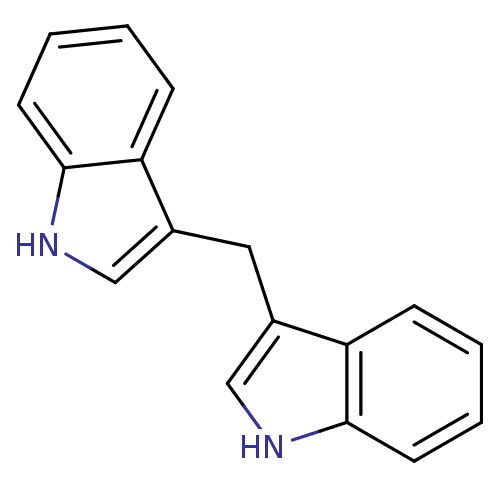 (1,1-Bis(3''-Indolyl)Methane | 3,3''-Diindolylmetha...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 7.60E+5 | n/a | n/a | n/a | n/a | n/a |
India Institute of Chemical Biology Curated by ChEMBL | Assay Description Binding affinity to recombinant drug-resistant Leishmania donovani LdDR50 topoisomerase 1b large subunit F270L, K430N mutant after 24 hrs by equilibr... | Antimicrob Agents Chemother 53: 2589-98 (2009) Article DOI: 10.1128/AAC.01648-08 BindingDB Entry DOI: 10.7270/Q2542RDS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA topoisomerase I (Leishmania donovani) | BDBM50239186 (1,1-Bis(3''-Indolyl)Methane | 3,3''-Diindolylmetha...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 5.30E+5 | n/a | n/a | n/a | n/a | n/a |
India Institute of Chemical Biology Curated by ChEMBL | Assay Description Binding affinity to recombinant drug-resistant Leishmania donovani LdDR50 topoisomerase 1b large subunit F270L mutant after 24 hrs by equilibrium dia... | Antimicrob Agents Chemother 53: 2589-98 (2009) Article DOI: 10.1128/AAC.01648-08 BindingDB Entry DOI: 10.7270/Q2542RDS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA topoisomerase type IB small subunit (Leishmania donovani) | BDBM50239186 (1,1-Bis(3''-Indolyl)Methane | 3,3''-Diindolylmetha...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 3.80E+5 | n/a | n/a | n/a | n/a | n/a |
India Institute of Chemical Biology Curated by ChEMBL | Assay Description Binding affinity to recombinant drug-resistant Leishmania donovani LdDR50 topoisomerase 1b small subunit N184S mutant after 24 hrs by equilibrium dia... | Antimicrob Agents Chemother 53: 2589-98 (2009) Article DOI: 10.1128/AAC.01648-08 BindingDB Entry DOI: 10.7270/Q2542RDS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||