

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Peroxisome proliferator-activated receptor gamma [182-505]/Regulatory protein GAL4 [1-147] | ||
| Ligand | BDBM222700 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Activation Assay | ||
| EC50 | 2740±n/a nM | ||
| Citation |  Miura, T; Sato, S; Yamada, H; Tagashira, J; Watanabe, T; Sekimoto, R; Ishida, R; Aoki, H; Ohgiya, T Phenylpyridine derivative and drug containing same US Patent US9315493 Publication Date 4/19/2016 Miura, T; Sato, S; Yamada, H; Tagashira, J; Watanabe, T; Sekimoto, R; Ishida, R; Aoki, H; Ohgiya, T Phenylpyridine derivative and drug containing same US Patent US9315493 Publication Date 4/19/2016 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Peroxisome proliferator-activated receptor gamma [182-505]/Regulatory protein GAL4 [1-147] | |||
| Name: | Peroxisome proliferator-activated receptor gamma [182-505]/Regulatory protein GAL4 [1-147] | ||
| Synonyms: | PPARgamma2 - DNA binding domain of Gal4 | ||
| Type: | DNA-Protein | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Regulatory protein GAL4 [1-147] | ||
| Synonyms: | DNA-binding transcription factor (GAl4) | GAL4 | GAL4_YEAST | ||
| Type: | DNA Binding | ||
| Mol. Mass.: | 16874.50 | ||
| Organism: | Saccharomyces bayanus (Yeast) | ||
| Description: | amino acids 1 to 147 | ||
| Residue: | 147 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | Peroxisome proliferator-activated receptor gamma [182-505] | ||
| Synonyms: | NR1C3 | PPARG | PPARG_HUMAN | Peroxisome proliferator-activated receptor gamma (PPAR-gamma) | ||
| Type: | Protein | ||
| Mol. Mass.: | 37077.80 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | amino acids 182 to 505 | ||
| Residue: | 324 | ||
| Sequence: |
| ||
| BDBM222700 | |||
| n/a | |||
| Name | BDBM222700 | ||
| Synonyms: | US9315493, 32 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H24ClF3N8O | ||
| Mol. Mass. | 580.991 | ||
| SMILES | CCCCc1nc(C)n(-c2ncc(cc2Cl)C(F)(F)F)c(=O)c1Cc1ccc(cn1)-c1ccccc1-c1nnn[nH]1 |(-7.34,.87,;-6,.1,;-4.67,.87,;-3.33,.1,;-2,.87,;-2,2.41,;-.67,3.18,;-.67,4.72,;.67,2.41,;2,3.18,;3.33,2.41,;4.67,3.18,;4.72,4.75,;3.33,5.49,;2,4.72,;.67,5.49,;6.05,5.52,;7.38,6.29,;6.82,4.19,;5.28,6.85,;.67,.87,;2,.1,;-.67,.1,;-.67,-1.44,;.67,-2.21,;2,-1.44,;3.33,-2.21,;3.33,-3.75,;2,-4.52,;.67,-3.75,;4.67,-4.52,;4.67,-6.06,;6,-6.83,;7.34,-6.06,;7.34,-4.52,;6,-3.75,;6,-2.21,;4.76,-1.3,;5.23,.16,;6.77,.16,;7.25,-1.3,)| | ||
| Structure | 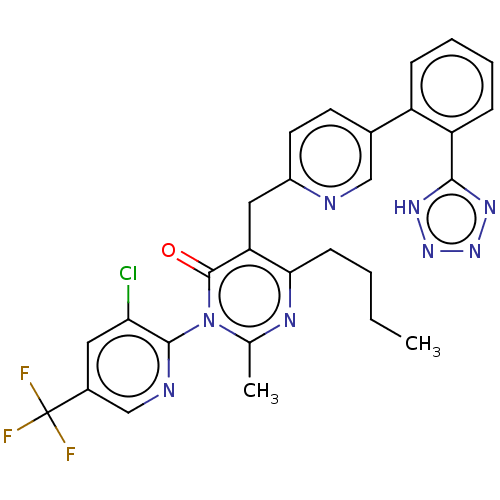
| ||