| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-dependent translocase ABCB1 bd-vinblastine |
|---|
| Ligand | BDBM227643 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Fluorescence Correlation Spectroscopy (FCS) |
|---|
| Temperature | 295.65±0.5 K |
|---|
| IC50 | 8.8e+3± 6e+3 nM |
|---|
| Citation |  Li, MJ; Nath, A; Atkins, WM Differential Coupling of Binding, ATP Hydrolysis, and Transport of Fluorescent Probes with P-Glycoprotein in Lipid Nanodiscs. Biochemistry56:2506-2517 (2017) [PubMed] Article Li, MJ; Nath, A; Atkins, WM Differential Coupling of Binding, ATP Hydrolysis, and Transport of Fluorescent Probes with P-Glycoprotein in Lipid Nanodiscs. Biochemistry56:2506-2517 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| ATP-dependent translocase ABCB1 bd-vinblastine |
|---|
| Name: | ATP-dependent translocase ABCB1 bd-vinblastine |
|---|
| Synonyms: | n/a |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | ATP-dependent translocase ABCB1 |
|---|
| Synonyms: | Abcb1a | Abcb4 | MDR1A_MOUSE | Mdr1a | Mdr3 | P-glycoprotein (P-gp) | P-glycoprotein 1 and 3 (MDR1a/MDR1b) | P-glycoprotein 3 | Pgy-3 | Pgy3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 140669.02 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | P21447 |
|---|
| Residue: | 1276 |
|---|
| Sequence: | MELEEDLKGRADKNFSKMGKKSKKEKKEKKPAVSVLTMFRYAGWLDRLYMLVGTLAAIIH
GVALPLMMLIFGDMTDSFASVGNVSKNSTNMSEADKRAMFAKLEEEMTTYAYYYTGIGAG
VLIVAYIQVSFWCLAAGRQIHKIRQKFFHAIMNQEIGWFDVHDVGELNTRLTDDVSKINE
GIGDKIGMFFQAMATFFGGFIIGFTRGWKLTLVILAISPVLGLSAGIWAKILSSFTDKEL
HAYAKAGAVAEEVLAAIRTVIAFGGQKKELERYNNNLEEAKRLGIKKAITANISMGAAFL
LIYASYALAFWYGTSLVISKEYSIGQVLTVFFSVLIGAFSVGQASPNIEAFANARGAAYE
VFKIIDNKPSIDSFSKSGHKPDNIQGNLEFKNIHFSYPSRKEVQILKGLNLKVKSGQTVA
LVGNSGCGKSTTVQLMQRLYDPLDGMVSIDGQDIRTINVRYLREIIGVVSQEPVLFATTI
AENIRYGREDVTMDEIEKAVKEANAYDFIMKLPHQFDTLVGERGAQLSGGQKQRIAIARA
LVRNPKILLLDEATSALDTESEAVVQAALDKAREGRTTIVIAHRLSTVRNADVIAGFDGG
VIVEQGNHDELMREKGIYFKLVMTQTAGNEIELGNEACKSKDEIDNLDMSSKDSGSSLIR
RRSTRKSICGPHDQDRKLSTKEALDEDVPPASFWRILKLNSTEWPYFVVGIFCAIINGGL
QPAFSVIFSKVVGVFTNGGPPETQRQNSNLFSLLFLILGIISFITFFLQGFTFGKAGEIL
TKRLRYMVFKSMLRQDVSWFDDPKNTTGALTTRLANDAAQVKGATGSRLAVIFQNIANLG
TGIIISLIYGWQLTLLLLAIVPIIAIAGVVEMKMLSGQALKDKKELEGSGKIATEAIENF
RTVVSLTREQKFETMYAQSLQIPYRNAMKKAHVFGITFSFTQAMMYFSYAACFRFGAYLV
TQQLMTFENVLLVFSAIVFGAMAVGQVSSFAPDYAKATVSASHIIRIIEKTPEIDSYSTQ
GLKPNMLEGNVQFSGVVFNYPTRPSIPVLQGLSLEVKKGQTLALVGSSGCGKSTVVQLLE
RFYDPMAGSVFLDGKEIKQLNVQWLRAQLGIVSQEPILFDCSIAENIAYGDNSRVVSYEE
IVRAAKEANIHQFIDSLPDKYNTRVGDKGTQLSGGQKQRIAIARALVRQPHILLLDEATS
ALDTESEKVVQEALDKAREGRTCIVIAHRLSTIQNADLIVVIQNGKVKEHGTHQQLLAQK
GIYFSMVSVQAGAKRS
|
|
|
|---|
| Component 2 |
| Name | BDBM227642 |
|---|
| Synonyms: | BD-vinblastine | BODIPY-vinblastine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C58H71BF2N6O9 |
|---|
| Mol. Mass. | 1045.027 |
|---|
| SMILES | CC[C@]1(O)CC2CN(C1)CCc1c3C=CCCc3[nH]c1[C@@](C2)(C(=O)OC)c1cc2c(cc1OC)N(C)[C@@H]1[C@]22CCN3CC=C[C@](CC)([C@@H]23)C(OC(=O)CCC2=[N+]3C(C=C2)=Cc2c(C)cc(C)n2[B-]3(F)F)[C@]1(O)C(=O)OC |r,c:14,48,65,67,t:62| |
|---|
| Structure | 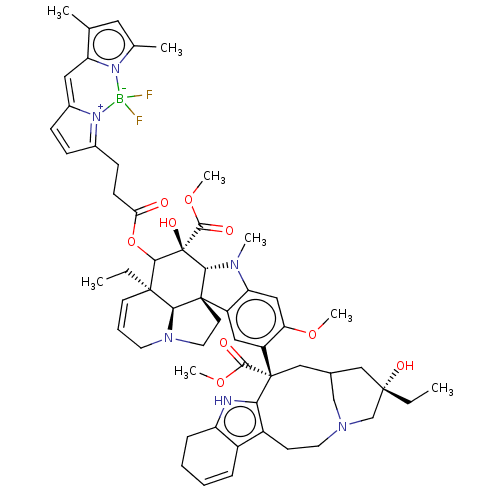
|
|---|
| BDBM227643 |
|---|
| n/a |
|---|
| Name | BDBM227643 |
|---|
| Synonyms: | Vinblastine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C46H58N4O9 |
|---|
| Mol. Mass. | 810.9741 |
|---|
| SMILES | CC[C@]1(O)C[C@H]2CN(C1)CCc1c([nH]c3ccccc13)[C@@](C2)(C(=O)OC)c1cc2c(cc1OC)N(C)[C@@H]1[C@]22CCN3CC=C[C@](CC)([C@@H]23)[C@@H](OC(C)=O)[C@]1(O)C(=O)OC |c:48| |
|---|
| Structure | 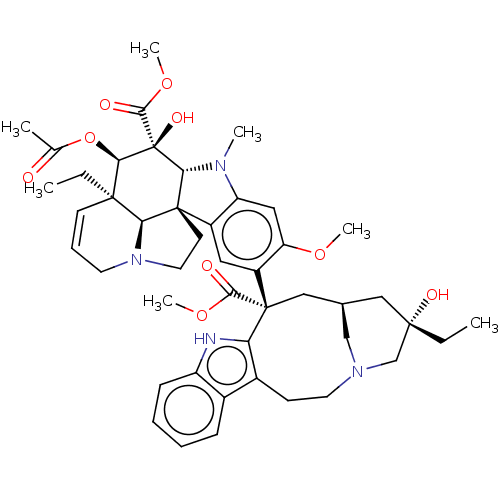
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, MJ; Nath, A; Atkins, WM Differential Coupling of Binding, ATP Hydrolysis, and Transport of Fluorescent Probes with P-Glycoprotein in Lipid Nanodiscs. Biochemistry56:2506-2517 (2017) [PubMed] Article
Li, MJ; Nath, A; Atkins, WM Differential Coupling of Binding, ATP Hydrolysis, and Transport of Fluorescent Probes with P-Glycoprotein in Lipid Nanodiscs. Biochemistry56:2506-2517 (2017) [PubMed] Article
