| Synonyms: | PI3-kinase p110 subunit delta | PI3-kinase p110-delta subunit | PI3-kinase subunit p110-delta | PI3K | PIK3CD | PK3CD_HUMAN | Phosphatidylinositol 4,5-biphosphate 3-kinase catalytic subunit delta (PIK3CD) | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit delta | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit delta (PI3K delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta (PI3Kdelta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform ( PI3K-delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3K delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3K-delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3Kd) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3Kdelta) |
|---|
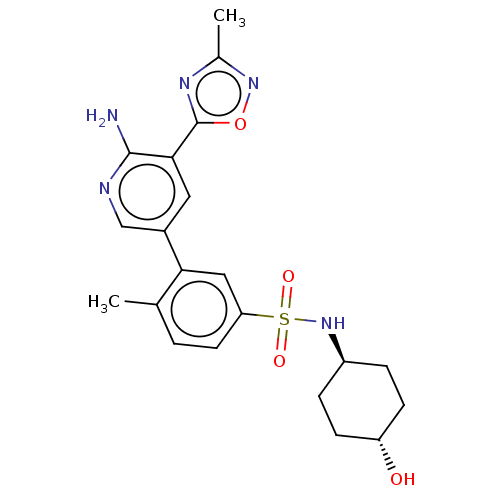
| SMILES | Cc1noc(n1)-c1cc(cnc1N)-c1cc(ccc1C)S(=O)(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:24.26,wD:27.30,(7.05,-.65,;6.28,.68,;7.19,1.93,;6.28,3.17,;4.82,2.69,;4.82,1.15,;3.48,3.47,;2.15,2.69,;.82,3.47,;.82,5,;2.15,5.78,;3.48,5,;4.82,5.78,;-.52,2.69,;-.52,1.15,;-1.85,.38,;-3.19,1.15,;-3.19,2.69,;-1.85,3.47,;-1.85,5,;-1.85,-1.15,;-.31,-1.15,;-3.39,-1.15,;-1.85,-2.69,;-3.19,-3.47,;-3.19,-5,;-4.52,-5.78,;-5.85,-5,;-7.19,-5.78,;-5.85,-3.47,;-4.52,-2.69,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bellenie, BR; Bloomfield, GC; Bruce, I; Culshaw, AJ; Hall, EC; Hollingworth, GJ; Neef, J; Spendiff, M; Watson, SJ Amino pyridine derivatives as phosphatidylinositol 3-kinase inhibitors US Patent US10112926 Publication Date 10/30/2018
Bellenie, BR; Bloomfield, GC; Bruce, I; Culshaw, AJ; Hall, EC; Hollingworth, GJ; Neef, J; Spendiff, M; Watson, SJ Amino pyridine derivatives as phosphatidylinositol 3-kinase inhibitors US Patent US10112926 Publication Date 10/30/2018