

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Ligand | BDBM451123 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Exemplary MCL-1 Inhibitors Bind MCL-1 | ||
| Ki | 0.023±n/a nM | ||
| Citation |  Judd, AS; Kunzer, AR; Lai, C; Souers, AJ; Tao, Z; Mastracchio, A; Wang, X; Ji, C; Wendt, MD; Song, X; Doherty, GA; Penning, TD Macrocyclic MCL-1 inhibitors and methods of use US Patent US10676485 Publication Date 6/9/2020 Judd, AS; Kunzer, AR; Lai, C; Souers, AJ; Tao, Z; Mastracchio, A; Wang, X; Ji, C; Wendt, MD; Song, X; Doherty, GA; Penning, TD Macrocyclic MCL-1 inhibitors and methods of use US Patent US10676485 Publication Date 6/9/2020 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Induced myeloid leukemia cell differentiation protein Mcl-1 | |||
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT | ||
| Type: | Membrane; Single-pass membrane protein | ||
| Mol. Mass.: | 37332.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q07820 | ||
| Residue: | 350 | ||
| Sequence: |
| ||
| BDBM451123 | |||
| n/a | |||
| Name | BDBM451123 | ||
| Synonyms: | US10676485, Example 33 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C53H61Cl2FN6O9S | ||
| Mol. Mass. | 1048.056 | ||
| SMILES | CN1CCN(C[C@@H]2COc3ccc(OCc4ccnc(n4)[C@H]4CC[C@](F)(COC[C@@H]5COCCO5)CC4)c(C[C@@H](Oc4ncnc5sc(C6=CCCC6)c(-c6c(C)c(Cl)c(O2)c(Cl)c6C)c45)C(O)=O)c3)CC1 |r,wU:39.73,6.5,24.26,wD:21.21,29.29,24.25,t:51,(15.65,6.11,;14.17,5.71,;13.77,4.22,;12.28,3.82,;11.19,4.91,;9.7,4.51,;9.31,3.03,;7.82,2.63,;4.67,5.47,;3.9,4.13,;2.36,4.13,;1.59,2.8,;2.36,1.47,;1.59,.13,;.05,.13,;-.72,1.47,;.05,2.8,;-.72,4.13,;-2.26,4.13,;-3.03,2.8,;-2.26,1.47,;-4.57,2.8,;-5.34,4.13,;-6.88,4.13,;-7.65,2.8,;-8.99,2.03,;-8.99,3.57,;-10.32,2.8,;-11.65,3.57,;-12.99,2.8,;-12.99,1.26,;-14.32,.49,;-15.65,1.26,;-15.65,2.8,;-14.32,3.57,;-6.88,1.47,;-5.34,1.47,;3.9,1.47,;4.67,.13,;4.67,-1.41,;6,-2.18,;6,-3.72,;4.67,-4.49,;4.67,-6.03,;6,-6.8,;7.34,-6.03,;8.8,-6.5,;9.71,-5.26,;11.25,-5.26,;12.15,-4.01,;13.62,-4.49,;13.62,-6.03,;12.15,-6.5,;8.8,-4.01,;9.2,-2.53,;8.11,-1.44,;6.62,-1.04,;8.51,.05,;7.42,1.14,;10,.45,;10.39,1.94,;11.08,-.64,;12.57,-.24,;10.69,-2.13,;11.78,-3.22,;7.34,-4.49,;3.33,-2.18,;2,-1.41,;3.33,-3.72,;4.67,2.8,;11.59,6.4,;13.08,6.8,)| | ||
| Structure | 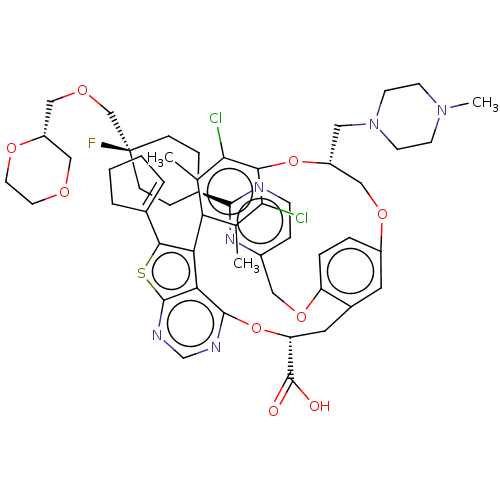
| ||