null
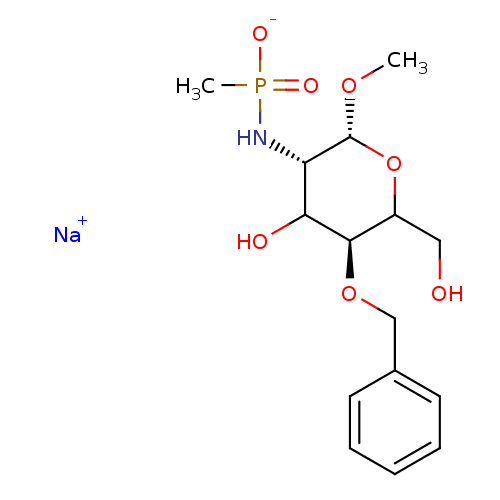
SMILES [Na+].CO[C@@H]1OC(CO)[C@@H](OCc2ccccc2)C(O)[C@@H]1NP(C)([O-])=O
InChI Key InChIKey=YFNJPBDPIGTYFU-FWJBWGTRSA-M
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 150805
Found 2 hits for monomerid = 150805
TargetPeptidoglycan-N-acetylglucosamine deacetylase(Streptococcus pneumoniae (Firmicutes))
University of Toronto
University of Toronto
Affinity DataKi: 1.70E+5nM ΔG°: -5.14kcal/molepH: 7.0 T: 2°CAssay Description:In a standard optimized reaction, SpPgdA was added to a microcentrifuge tube and incubated at 30 °C for 5 min. Chitotriose was added as a substrate t...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataKi: >1.00E+6nMpH: 7.5Assay Description:Reactions were carried out in solutions (50 µL) containing PgaB (10 µM) in HEPES (100 mM, pH 7.5), and varying concentrations of ACC (0.05-10 mM) at ...More data for this Ligand-Target Pair