null
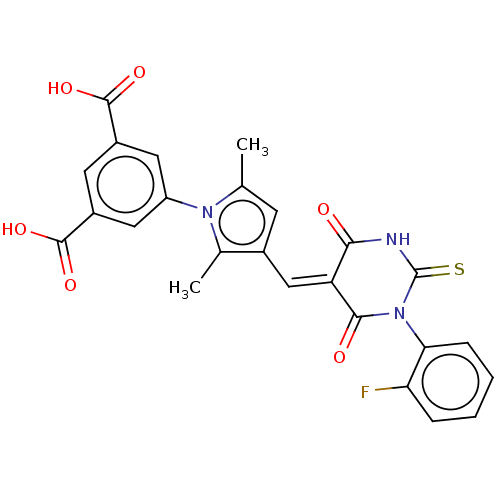
SMILES Cc1cc(\C=C2\C(=O)NC(=S)N(C2=O)c2ccccc2F)c(C)n1-c1cc(cc(c1)C(O)=O)C(O)=O
InChI Key InChIKey=ANNMFYDZXMVBCO-WQRHYEAKSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50046823
Found 2 hits for monomerid = 50046823
TargetUDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase(Escherichia coli UMNK88 (Enterobacteria))
Freie Universit£t Berlin
Curated by ChEMBL
Freie Universit£t Berlin
Curated by ChEMBL
Affinity DataIC50: 3.11E+5nMAssay Description:Inhibition of Escherichia coli MurE after 15 mins in presence of 0.005% Triton X-114 by Malachite green assayMore data for this Ligand-Target Pair
TargetUDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase(Escherichia coli)
Freie Universit£t Berlin
Curated by ChEMBL
Freie Universit£t Berlin
Curated by ChEMBL
Affinity DataIC50: 3.44E+5nMAssay Description:Inhibition of Escherichia coli MurF after 15 mins in presence of 0.005% Triton X-114 by Malachite green assayMore data for this Ligand-Target Pair