null
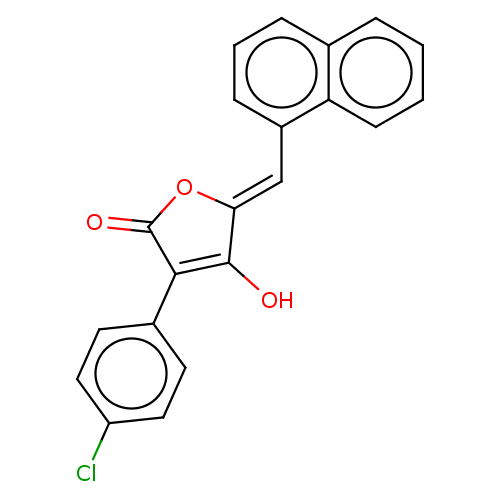
SMILES OC1=C(C(=O)O\C1=C/c1cccc2ccccc12)c1ccc(Cl)cc1
InChI Key InChIKey=PLGHLEBIWUQEPR-PDGQHHTCSA-N
PDB links: 1 PDB ID matches this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50475588
Found 4 hits for monomerid = 50475588
TargetUDP-N-acetylmuramoylalanine--D-glutamate ligase(Escherichia coli (strain K12))
Wyeth Research
Curated by ChEMBL
Wyeth Research
Curated by ChEMBL
Affinity DataIC50: 7.16E+4nMAssay Description:Inhibitory activity against MurD in Escherichia coliMore data for this Ligand-Target Pair
TargetUDP-N-acetylenolpyruvoylglucosamine reductase(Escherichia coli K-12 (Enterobacteria))
Wyeth Research
Curated by ChEMBL
Wyeth Research
Curated by ChEMBL
Affinity DataIC50: 2.86E+4nMAssay Description:Inhibitory activity against MurB in Escherichia coliMore data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+4nMAssay Description:Inhibitory activity against MurC in Escherichia coliMore data for this Ligand-Target Pair
TargetUDP-N-acetylglucosamine 1-carboxyvinyltransferase(Escherichia coli K-12 (Enterobacteria))
Wyeth Research
Curated by ChEMBL
Wyeth Research
Curated by ChEMBL
Affinity DataIC50: 7.16E+4nMAssay Description:Inhibitory activity against MurA in Escherichia coliMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)